Welcome to the Nanopore Community
Order MinION devices and consumables
Visit vwr.comPCR tiling of SARS-CoV-2 virus - Rapid Barcoding Kit 96 V14 and Midnight RT PCR Expansion (SQK-RBK114.96 and EXP-MRT001)
Version for device: MinION
Introduction to the protocol
Overview of the protocol
Overview of the protocol
-
Introduction to the protocol
To enable support for the rapidly expanding user requests, the team at Oxford Nanopore Technologies have put together an updated workflow based on the ARTIC Network protocols and analysis methods. The protocol uses Oxford Nanopore Technologies' Rapid Barcoding Kit 96 V14 (SQK-RBK114.96) and Midnight RT PCR Expansion (EXP-MRT001) for barcoding and library preparation.
While this protocol is available in the Nanopore Community, we kindly ask users to ensure they are citing the members of the ARTIC network who have been behind the development of these methods.
This protocol is similar to the ARTIC amplicon sequencing protocol for MinION for SARS-CoV-2 v3 (LoCost) by Josh Quick and the method used in Freed et al., 2020. The protocol generates amplicons in a tiled fashion across the whole SARS-CoV-2 genome.
To generate tiled PCR amplicons from the SARS-CoV-2 viral cDNA for use with the Rapid Barcoding Kit 96 V14 (SQK-RBK114.96), primers were designed by Freed et al., 2020 using Primal Scheme. These primers are in the Midnight RT PCR Expansion (EXP-MRT001) and are designed to generate 1.2 kb amplicons. Primer sequences can be found here.
As mutations in SARS-CoV-2 variants emerge amplicon drop out may be observed; for users wishing to design their own primer spike-ins to address this we suggest adding to the appropriate primer pool at a final concentration between 3.33 µM and 6.66 µM.
Steps in the sequencing workflow:
Prepare for your experiment
you will need to:- Extract your RNA
- Ensure you have your sequencing kit, the correct equipment and reagents
- Download the software for acquiring and analysing your data
- Check your flow cell to ensure it has enough pores for a good sequencing run
Prepare your library
You will need to:- Reverse transcribe your RNA samples with random hexamers
- Amplify the samples by tiled PCR using separate primer pools
- Combine the primer pools
- Attach Rapid Barcodes supplied in the kit to the DNA ends, pool the samples and SPRI purify
- Prime the flow cell and load your DNA library into the flow cell
Sequencing and analysis
You will need to:- Start a sequencing run using the MinKNOW software, selecting SQK-RBK114.96 in kit selection, which will collect raw data from the device and convert it into basecalled reads
- (Optional): Perform downstream analysis of the data using the wf-artic analysis workflow integrated within the EPI2ME Labs application
-
Before starting
This protocol outlines how to carry out PCR tiling of SARS-CoV-2 viral RNA samples on a 96-well plate using the Rapid Barcoding Kit 96 V14 (SQK-RBK114.96) with the Midnight RT PCR Expansion (EXP-MRT001).
It is required to use total RNA extracted from samples that have been screened by a suitable qPCR assay.
When processing multiple samples at once, we recommend making master mixes with an additional 10% of the volume. We also recommend using a template-free pre-PCR hood for making up the master mixes, and a separate template pre-PCR hood for handling the samples. It is important to clean and/or UV irradiate these hoods between sample batches. Furthermore, to track and monitor cross-contamination events, it is important to run a negative control reaction at the reverse transcription stage using nuclease-free water instead of sample, and carrying this control through the rest of the prep.
All post-PCR procedures must be carried out in a separate area to the pre-PCR preparation, with dedicated equipment for liquid handling in each area.
Equipment and consumables
- Materials
-
- Input RNA in 10 mM Tris-HCl, pH 8.0
- Rapid Barcoding Kit 96 V14 (SQK-RBK114.96)
- Midnight RT PCR Expansion (EXP-MRT001)
- Consumables
-
- Nuclease-free water (e.g. ThermoFisher, AM9937)
- Freshly prepared 80% ethanol in nuclease-free water
- Qubit dsDNA HS Assay Kit (Invitrogen, Q32851)
- Qubit™ Assay Tubes (Invitrogen, Q32856)
- 1.5 ml Eppendorf DNA LoBind tubes
- 2 ml Eppendorf DNA LoBind tubes
- 5 ml Eppendorf DNA LoBind tubes
- Eppendorf twin.tec® PCR plate 96 LoBind, semi-skirted (Cat # 0030129504) with PCR seals
- Bovine Serum Albumin (BSA) (50 mg/ml) (e.g Invitrogen™ UltraPure™ BSA 50 mg/ml, AM2616)
- Equipment
-
- Hula mixer (gentle rotator mixer)
- Magnetic rack
- Centrifuge capable of taking 96-well plates
- Microfuge
- Vortex mixer
- Thermal cycler
- Multichannel pipettes suitable for dispensing 0.5–10 μl, 2–20 μl and 20–200 μl, and tips
- P1000 pipette and tips
- P200 pipette and tips
- P100 pipette and tips
- P20 pipette and tips
- P10 pipette and tips
- Ice bucket with ice
- Timer
- Qubit fluorometer (or equivalent)
- Optional Equipment
-
- Eppendorf 5424 centrifuge (or equivalent)
- PCR hood with UV steriliser (optional but recommended to reduce cross-contamination)
- PCR-Cooler (Eppendorf)
- Stepper pipette and tips
-
For this protocol, you will need your extracted RNA in 8 µl 10 mM Tris-HCl, pH 8.0.
-
Rapid Barcoding Kit 96 V14 (SQK-RBK114.96) contents
Name Acronym Cap colour No. of vials Fill volume per vial (µl) Rapid Adapter RA Green 2 15 Adapter Buffer ADB Clear 1 100 AMPure XP Beads AXP Amber 3 1,200 Elution Buffer EB Black 1 1,500 Sequencing Buffer SB Red 1 1,700 Library Beads LIB Pink 1 1,800 Library Solution LIS White cap, pink label 1 1,800 Flow Cell Flush FCF Clear 1 15,500 Flow Cell Tether FCT Purple 2 200 Rapid Barcodes RB01-96 - 3 plates 8 µl per well This Product Contains AMPure XP Reagent Manufactured by Beckman Coulter, Inc. and can be stored at -20°C with the kit without detriment to reagent stability.
-
Midnight RT PCR Expansion (EXP-MRT001) contents
Name Acronym Cap colour Number of vials Fill volume per vial (µl) LunaScript RT SuperMix LS RT Blue 3 500 Q5 HS Master Mix Q5 Orange 6 1,500 Midnight Primer Pool A MP A White 3 15 Midnight Primer Pool B MP B Clear 3 15 -
Midnight Primer sequences
As mutations in SARS-CoV-2 variants emerge amplicon drop out may be observed; for users wishing to design their own primer spike-ins to address this we suggest adding to the appropriate primer pool at a final concentration between 3.33 µM and 6.66 µM.
Below are the sequences for the V3 primer scheme used in the Midnight RT PCR Expansion.
Pool A
Primer name Primer Sequence SARSCoV_1200_1_LEFTACCAACCAACTTTCGATCTCTTGT SARSCoV_1200_1_RIGHTGGTTGCATTCATTTGGTGACGC SARSCoV_1200_3_LEFTGGCTTGAAGAGAAGTTTAAGGAAGGT SARSCoV_1200_3_RIGHTGATTGTCCTCACTGCCGTCTTG SARSCoV_1200_5_LEFTACCTACTAAAAAGGCTGGTGGC SARSCoV_1200_5_RIGHTAGCATCTTGTAGAGCAGGTGGA SARSCoV_1200_7_LEFTACCTGGTGTATACGTTGTCTTTGG SARSCoV_1200_7_RIGHTGCTGAAATCGGGGCCATTTGTA SARSCoV_1200_9_LEFTAGAAGTTACTGGCGATAGTTGTAATAACT SARSCoV_1200_9_RIGHTTGCTGATATGTCCAAAGCACCA SARSCoV_1200_11_LEFTAGACACCTAAGTATAAGTTTGTTCGCA SARSCoV_1200_11_RIGHTGCCCACATGGAAATGGCTTGAT SARSCoV_1200_13_LEFTACCTCTTACAACAGCAGCCAAAC SARSCoV_1200_13_RIGHTCGTCCTTTTCTTGGAAGCGACA SARSCoV_1200_15_LEFTTTTTAAGGAATTACTTGTGTATGCTGCT SARSCoV_1200_15_RIGHTACACACAACAGCATCGTCAGAG SARSCoV_1200_17_LEFTTCAAGCTTTTTGCAGCAGAAACG SARSCoV_1200_17_RIGHTCCAAGCAGGGTTACGTGTAAGG SARSCoV_1200_19_LEFTGGCACATGGCTTTGAGTTGACA SARSCoV_1200_19_RIGHTCCTGTTGTCCATCAAAGTGTCCC SARSCoV_1200_21_LEFTTCTGTAGTTTCTAAGGTTGTCAAAGTGA SARSCoV_1200_21_RIGHTGCAGGGGGTAATTGAGTTCTGG 21_right_spikeGTGTATGATTGAGTTCTGGTTGTAAG SARSCoV_1200_23_LEFTACTTTAGAGTCCAACCAACAGAATCT 23_left_spikeACTTTAGAGTTCAACCAACAGAATCT SARSCoV_1200_23_RIGHTTGACTAGCTACACTACGTGCCC SARSCoV_1200_25_LEFTTGCTGCTACTAAAATGTCAGAGTGT SARSCoV_1200_25_RIGHTCATTTCCAGCAAAGCCAAAGCC SARSCoV_1200_27_LEFTTGGATCACCGGTGGAATTGCTA SARSCoV_1200_27_RIGHTTGTTCGTTTAGGCGTGACAAGT SARSCoV_1200_29_LEFTTGAGGGAGCCTTGAATACACCA SARSCoV_1200_29_RIGHTTAGGCAGCTCTCCCTAGCATTG Pool B
Primer name Primer sequences SARSCoV_1200_2_LEFTCCATAATCAAGACTATTCAACCAAGGGT SARSCoV_1200_2_RIGHTACAGGTGACAATTTGTCCACCG SARSCoV_1200_4_LEFTGGAATTTGGTGCCACTTCTGCT SARSCoV_1200_4_RIGHTCCTGACCCGGGTAAGTGGTTAT SARSCoV_1200_6_LEFTACTTCTATTAAATGGGCAGATAACAACTG SARSCoV_1200_6_RIGHTGATTATCCATTCCCTGCGCGTC SARSCoV_1200_8_LEFTCAATCATGCAATTGTTTTTCAGCTATTTTG SARSCoV_1200_8_RIGHTTGACTTTTTGCTACCTGCGCAT SARSCoV_1200_10_LEFTTTTACCAGGAGTTTTCTGTGGTGT SARSCoV_1200_10_RIGHTTGGGCCTCATAGCACATTGGTA SARSCoV_1200_12_LEFTATGGTGCTAGGAGAGTGTGGAC SARSCoV_1200_12_RIGHTGGATTTCCCACAATGCTGATGC SARSCoV_1200_14_LEFTACAGGCACTAGTACTGATGTCGT SARSCoV_1200_14_RIGHTGTGCAGCTACTGAAAAGCACGT SARSCoV_1200_16_LEFTACAACACAGACTTTATGAGTGTCTCT SARSCoV_1200_16_RIGHTCTCTGTCAGACAGCACTTCACG SARSCoV_1200_18_LEFTGCACATAAAGACAAATCAGCTCAATGC SARSCoV_1200_18_RIGHTTGTCTGAAGCAGTGGAAAAGCA SARSCoV_1200_20_LEFTACAATTTGATACTTATAACCTCTGGAACAC SARSCoV_1200_20_RIGHTGATTAGGCATAGCAACACCCGG SARSCoV_1200_22_LEFTGTGATGTTCTTGTTAACAACTAAACGAACA SARSCoV_1200_22_RIGHTAACAGATGCAAATCTGGTGGCG 22_right_spikeAACAGATGCAAATTTGGTGGCG SARSCoV_1200_24_LEFTGCTGAACATGTCAACAACTCATATGA 24_left_spikeGCTGAATATGTCAACAACTCATATGA SARSCoV_1200_24_RIGHTATGAGGTGCTGACTGAGGGAAG SARSCoV_1200_26_LEFTGCCTTGAAGCCCCTTTTCTCTA SARSCoV_1200_26_RIGHTAATGACCACATGGAACGCGTAC SARSCoV_1200_28_LEFTTTTGTGCTTTTTAGCCTTTCTGCT SARSCoV_1200_28_RIGHTGTTTGGCCTTGTTGTTGTTGGC SARSCoV_1200_28_LEFT_27837TTTTGTGCTTTTTAGCCTTTCTGTT -
Rapid barcode sequences
Component Sequence RB01 AAGAAAGTTGTCGGTGTCTTTGTG RB02 TCGATTCCGTTTGTAGTCGTCTGT RB03 GAGTCTTGTGTCCCAGTTACCAGG RB04 TTCGGATTCTATCGTGTTTCCCTA RB05 CTTGTCCAGGGTTTGTGTAACCTT RB06 TTCTCGCAAAGGCAGAAAGTAGTC RB07 GTGTTACCGTGGGAATGAATCCTT RB08 TTCAGGGAACAAACCAAGTTACGT RB09 AACTAGGCACAGCGAGTCTTGGTT RB10 AAGCGTTGAAACCTTTGTCCTCTC RB11 GTTTCATCTATCGGAGGGAATGGA RB12 CAGGTAGAAAGAAGCAGAATCGGA RB13 AGAACGACTTCCATACTCGTGTGA RB14 AACGAGTCTCTTGGGACCCATAGA RB15 AGGTCTACCTCGCTAACACCACTG RB16 CGTCAACTGACAGTGGTTCGTACT RB17 ACCCTCCAGGAAAGTACCTCTGAT RB18 CCAAACCCAACAACCTAGATAGGC RB19 GTTCCTCGTGCAGTGTCAAGAGAT RB20 TTGCGTCCTGTTACGAGAACTCAT RB21 GAGCCTCTCATTGTCCGTTCTCTA RB22 ACCACTGCCATGTATCAAAGTACG RB23 CTTACTACCCAGTGAACCTCCTCG RB24 GCATAGTTCTGCATGATGGGTTAG RB25 GTAAGTTGGGTATGCAACGCAATG RB26 CATACAGCGACTACGCATTCTCAT RB27 CGACGGTTAGATTCACCTCTTACA RB28 TGAAACCTAAGAAGGCACCGTATC RB29 CTAGACACCTTGGGTTGACAGACC RB30 TCAGTGAGGATCTACTTCGACCCA RB31 TGCGTACAGCAATCAGTTACATTG RB32 CCAGTAGAAGTCCGACAACGTCAT RB33 CAGACTTGGTACGGTTGGGTAACT RB34 GGACGAAGAACTCAAGTCAAAGGC RB35 CTACTTACGAAGCTGAGGGACTGC RB36 ATGTCCCAGTTAGAGGAGGAAACA RB37 GCTTGCGATTGATGCTTAGTATCA RB38 ACCACAGGAGGACGATACAGAGAA RB39 CCACAGTGTCAACTAGAGCCTCTC RB40 TAGTTTGGATGACCAAGGATAGCC RB41 GGAGTTCGTCCAGAGAAGTACACG RB42 CTACGTGTAAGGCATACCTGCCAG RB43 CTTTCGTTGTTGACTCGACGGTAG RB44 AGTAGAAAGGGTTCCTTCCCACTC RB45 GATCCAACAGAGATGCCTTCAGTG RB46 GCTGTGTTCCACTTCATTCTCCTG RB47 GTGCAACTTTCCCACAGGTAGTTC RB48 CATCTGGAACGTGGTACACCTGTA RB49 ACTGGTGCAGCTTTGAACATCTAG RB50 ATGGACTTTGGTAACTTCCTGCGT RB51 GTTGAATGAGCCTACTGGGTCCTC RB52 TGAGAGACAAGATTGTTCGTGGAC RB53 AGATTCAGACCGTCTCATGCAAAG RB54 CAAGAGCTTTGACTAAGGAGCATG RB55 TGGAAGATGAGACCCTGATCTACG RB56 TCACTACTCAACAGGTGGCATGAA RB57 GCTAGGTCAATCTCCTTCGGAAGT RB58 CAGGTTACTCCTCCGTGAGTCTGA RB59 TCAATCAAGAAGGGAAAGCAAGGT RB60 CATGTTCAACCAAGGCTTCTATGG RB61 AGAGGGTACTATGTGCCTCAGCAC RB62 CACCCACACTTACTTCAGGACGTA RB63 TTCTGAAGTTCCTGGGTCTTGAAC RB64 GACAGACACCGTTCATCGACTTTC RB65 TTCTCAGTCTTCCTCCAGACAAGG RB66 CCGATCCTTGTGGCTTCTAACTTC RB67 GTTTGTCATACTCGTGTGCTCACC RB68 GAATCTAAGCAAACACGAAGGTGG RB69 TACAGTCCGAGCCTCATGTGATCT RB70 ACCGAGATCCTACGAATGGAGTGT RB71 CCTGGGAGCATCAGGTAGTAACAG RB72 TAGCTGACTGTCTTCCATACCGAC RB73 AAGAAACAGGATGACAGAACCCTC RB74 TACAAGCATCCCAACACTTCCACT RB75 GACCATTGTGATGAACCCTGTTGT RB76 ATGCTTGTTACATCAACCCTGGAC RB77 CGACCTGTTTCTCAGGGATACAAC RB78 AACAACCGAACCTTTGAATCAGAA RB79 TCTCGGAGATAGTTCTCACTGCTG RB80 CGGATGAACATAGGATAGCGATTC RB81 CCTCATCTTGTGAAGTTGTTTCGG RB82 ACGGTATGTCGAGTTCCAGGACTA RB83 TGGCTTGATCTAGGTAAGGTCGAA RB84 GTAGTGGACCTAGAACCTGTGCCA RB85 AACGGAGGAGTTAGTTGGATGATC RB86 AGGTGATCCCAACAAGCGTAAGTA RB87 TACATGCTCCTGTTGTTAGGGAGG RB88 TCTTCTACTACCGATCCGAAGCAG RB89 ACAGCATCAATGTTTGGCTAGTTG RB90 GATGTAGAGGGTACGGTTTGAGGC RB91 GGCTCCATAGGAACTCACGCTACT RB92 TTGTGAGTGGAAAGATACAGGACC RB93 AGTTTCCATCACTTCAGACTTGGG RB94 GATTGTCCTCAAACTGCCACCTAC RB95 CCTGTCTGGAAGAAGAATGGACTT RB96 CTGAACGGTCATAGAGTCCACCAT
Computer requirements and software
Computer requirements and software
-
MinION Mk1B IT requirements
Sequencing on a MinION Mk1B requires a high-spec computer or laptop to keep up with the rate of data acquisition. For more information, refer to the MinION Mk1B IT requirements document.
-
MinION Mk1C IT requirements
The MinION Mk1C contains fully-integrated compute and screen, removing the need for any accessories to generate and analyse nanopore data. For more information refer to the MinION Mk1C IT requirements document.
-
MinION Mk1D IT requirements
Sequencing on a MinION Mk1D requires a high-spec computer or laptop to keep up with the rate of data acquisition. For more information, refer to the MinION Mk1D IT requirements document.
-
Software for nanopore sequencing
MinKNOW
The MinKNOW software controls the nanopore sequencing device, collects sequencing data and basecalls in real time. You will be using MinKNOW for every sequencing experiment to sequence, basecall and demultiplex if your samples were barcoded.
For instructions on how to run the MinKNOW software, please refer to the MinKNOW protocol.
EPI2ME (optional)
The EPI2ME cloud-based platform performs further analysis of basecalled data, for example alignment to the Lambda genome, barcoding, or taxonomic classification. You will use the EPI2ME platform only if you would like further analysis of your data post-basecalling.
For instructions on how to create an EPI2ME account and install the EPI2ME Desktop Agent, please refer to this link.
-
Check your flow cell
We highly recommend that you check the number of pores in your flow cell prior to starting a sequencing experiment. This should be done within 12 weeks of purchasing for MinION/GridION/PromethION or within four weeks of purchasing Flongle Flow Cells. Oxford Nanopore Technologies will replace any flow cell with fewer than the number of pores in the table below, when the result is reported within two days of performing the flow cell check, and when the storage recommendations have been followed. To do the flow cell check, please follow the instructions in the Flow Cell Check document.
Flow cell Minimum number of active pores covered by warranty Flongle Flow Cell 50 MinION/GridION Flow Cell 800 PromethION Flow Cell 5000
Library preparation
Reverse transcription
Reverse transcription
- Materials
-
- Input RNA in 10 mM Tris-HCl, pH 8.0
- LunaScript RT SuperMix (LS RT)
- Consumables
-
- Nuclease-free water (e.g. ThermoFisher, AM9937)
- Eppendorf twin.tec® PCR plate 96 LoBind, semi-skirted (Cat # 0030129504) with PCR seals
- Equipment
-
- Multichannel pipettes suitable for dispensing 0.5–10 μl, 2–20 μl and 20–200 μl, and tips
- Thermal cycler
- Centrifuge capable of taking 96-well plates
- Ice bucket with ice
- Optional Equipment
-
- PCR-Cooler (Eppendorf)
- PCR hood with UV steriliser (optional but recommended to reduce cross-contamination)
- Stepper pipette and tips
-
In a clean pre-PCR hood, place a fresh 96-well plate (RT plate) into a PCR Cooler (if using). Using a stepper pipette, or multichannel pipette, add 2 µl of LunaScript RT SuperMix (LS RT) per well.
Depending on the number of samples, fill each well per column as follows:
Plate location X24 samples X48 samples X96 samples Columns 1-3 1-6 1-12 -
To each well containing LunaScript RT SuperMix (LS RT), add 8 µl of sample and gently mix by pipetting. If adding less than 8 µl, make up the rest of the volume with nuclease-free water.
Example for X48 samples:
-
Seal the RT plate and spin down.
-
Incubate the samples in the thermal cycler using the following program:
Step Temperature Time Cycles Primer annealing 25°C 2 min 1 cDNA synthesis 55°C 10 min 1 Heat inactivation 95°C 1 min 1 Hold 4°C ∞
PCR
PCR
- Materials
-
- Q5 HS Master Mix (Q5)
- Midnight Primer Pool A (MP A)
- Midnight Primer Pool B (MP B)
- Consumables
-
- Nuclease-free water (e.g. ThermoFisher, AM9937)
- 1.5 ml Eppendorf DNA LoBind tubes
- Eppendorf twin.tec® PCR plate 96 LoBind, semi-skirted (Cat # 0030129504) with PCR seals
- Equipment
-
- Multichannel pipettes suitable for dispensing 0.5–10 μl, 2–20 μl and 20–200 μl, and tips
- P1000 pipette and tips
- P200 pipette and tips
- Thermal cycler
- Microfuge
- Centrifuge capable of taking 96-well plates
- Ice bucket with ice
- Optional Equipment
-
- PCR-Cooler (Eppendorf)
- PCR hood with UV steriliser (optional but recommended to reduce cross-contamination)
- Stepper pipette and tips
-
Primer design
To generate tiled PCR amplicons from the SARS-CoV-2 viral cDNA, primers were designed by Freed et al., 2020 using Primal Scheme. These primers are designed to generate 1200 bp amplicons that overlap by approximately 20 bp. These primer sequences can be found here.
-
In the template-free pre-PCR hood, prepare the following master mixes in Eppendorf DNA LoBind tubes and mix thoroughly as follows:
Volume per sample:
Reagent Pool A Pool B Nuclease-free water 3.7 µl 3.7 µl Midnight Primer Pool A (MP A) 0.05 µl - Midnight Primer Pool B (MP B) - 0.05 µl Q5 HS Master Mix (Q5) 6.25 µl 6.25 µl Total 10 µl 10 µl For x24 samples:
Reagent Pool A Pool B Nuclease-free water 102 µl 102 µl Midnight Primer Pool A (MP A) 2 µl - Midnight Primer Pool B (MP B) - 2 µl Q5 HS Master Mix (Q5) 172 µl 172 µl Total 276 µl 276 µl For x48 samples:
Reagent Pool A Pool B Nuclease-free water 203 µl 203 µl Midnight Primer Pool A (MP A) 3 µl - Midnight Primer Pool B (MP B) - 3 µl Q5 HS Master Mix (Q5) 344 µl 344 µl Total 550 µl 550 µl For x96 samples:
Reagent Pool A Pool B Nuclease-free water 407 µl 407 µl Midnight Primer Pool A (MP A) 6 µl - Midnight Primer Pool B (MP B) - 6 µl Q5 HS Master Mix (Q5) 687 µl 687 µl Total 1,100 µl 1,100 µl -
Using a stepper pipette or a multichannel pipette, aliquot 10 µl of Pool A and Pool B into a clean 96-well plate(s) as follows:
Plate location X24 samples X48 samples X96 samples Columns Pool A: 1-3
Pool B: 4-6Pool A: 1-6
Pool B: 7-12Pool A: 1-12
Pool B: 1-12Note: For X96 samples, Pool A is a separate plate to Pool B.
-
Using a multichannel pipette, transfer 2.5 μl of each RT reaction from the RT plate to the corresponding well for both Pool A and Pool B in the PCR plate(s), taking care not to cross-contaminate different wells. Mix by pipetting the contents of each well up and down.
There should be two PCR reactions per sample.
Example for X48 samples:
-
Mix by pipetting the contents of each well up and down.
-
Seal the plate(s) and spin down briefly.
-
Incubate using the following program, with the heated lid set to 105°C:
Step Temperature Time Cycles Initial denaturation 98°C 30 sec 1 Denaturation
Annealing and extension98°C
61°C
65°C15 sec
2 min
3 min
35Hold 4°C ∞ -
Optional ActionIf necessary, the protocol can be paused at this point. The samples should be kept at 4°C and can be stored overnight.
Addition of rapid barcodes
Addition of rapid barcodes
- Materials
-
- Rapid Barcode Plate (RB01-96)
- Consumables
-
- Nuclease-free water (e.g. ThermoFisher, AM9937)
- Eppendorf twin.tec® PCR plate 96 LoBind, semi-skirted (Cat # 0030129504) with PCR seals
- Equipment
-
- Multichannel pipettes suitable for dispensing 2–20 μl and 20–200 μl, and tips
- Thermal cycler
- Centrifuge capable of taking 96-well plates
-
Spin down the Rapid Barcode Plate and PCR reactions prior to opening to collect material in the bottom of the wells.
-
Using a multichannel pipette or stepper pipette, transfer 2.5 μl nuclease-free water to the wells of a fresh 96-well plate (Barcode Attachment Plate).
Depending on the number of samples, aliquot into each well of the columns as follows:
Plate location X24 samples X48 samples X96 samples Columns 1-3 1-6 1-12 -
Using a multichannel pipette, transfer the entire contents of each well of PCR Pool B to the corresponding well of PCR Pool A and mix by pipetting.
Depending on the number of samples, Pool B columns will correspond to different Pool A columns.
No. of samples Pool B column Corresponding Pool A column X24 4
5
61
2
3X48 7
8
9
10
11
121
2
3
4
5
6X96 1
2
3
4
5
6
7
8
9
10
11
121
2
3
4
5
6
7
8
9
10
11
12Example for X48 samples:
-
Using a multichannel pipette, transfer 5 µl from each well of PCR Pool A (now containing pooled PCR products) to the corresponding well of the Barcode Attachment Plate and mix by pipetting.
Depending on the number of samples, PCR Pool A will be in each well of the following columns:
Plate location X24 samples X48 samples X96 samples Columns 1-3 1-6 1-12 Example for X48 samples:
-
Using a multichannel pipette, transfer 2.5 μl from the Rapid Barcode Plate to the corresponding well of the Barcode Attachment Plate, taking care not to cross-contaminate different wells. Mix by pipetting.
Depending on the number of samples, aliquot into each well of the columns as follows:
Plate location X24 samples X48 samples X96 samples Columns 1-3 1-6 1-12 Example for X48 samples:
-
Seal the Barcode Attachment Plate and spin down.
-
Incubate the plate in a thermal cycler at 30°C for 2 minutes and then at 80°C for 2 minutes.
Pooling samples and clean-up
Pooling samples and clean-up
- Materials
-
- AMPure XP Beads (AXP)
- Elution Buffer from the Oxford Nanopore kit (EB)
- Rapid Adapter (RA)
- Adapter Buffer (ADB)
- Consumables
-
- Freshly prepared 80% ethanol in nuclease-free water
- 1.5 ml Eppendorf DNA LoBind tubes
- 5 ml Eppendorf DNA LoBind tubes
- Qubit dsDNA HS Assay Kit (Invitrogen, Q32851)
- Qubit™ Assay Tubes (Invitrogen, Q32856)
- Equipment
-
- Microfuge
- Centrifuge capable of taking 96-well plates
- Hula mixer (gentle rotator mixer)
- Magnetic rack
- Ice bucket with ice
- P1000 pipette and tips
- P200 pipette and tips
- P20 pipette and tips
- P10 pipette and tips
- Qubit fluorometer plate reader (or equivalent for QC check)
-
Briefly spin down the Barcode Attachment Plate to collect the liquid at the bottom of the wells prior to opening.
-
Pool the barcoded samples in a 1.5 ml Eppendorf DNA LoBind tube.
We expect to have about ~10 µl per sample.
X24 samples X48 samples X96 samples Total volume ~240 µl ~480 µl ~960 µl -
Mix pooled samples by vortexing.
-
Transfer half of the barcoded pooled sample to a clean 1.5 ml Eppendorf DNA LoBind tube.
Per sample, we expect to take forward ~5 µl.
X24 samples X48 samples X96 samples Example volume 120 µl 240 µl 480 µl -
Resuspend the AMPure XP Beads (AXP) by vortexing.
-
To the pooled barcoded sample, add an equal volume of resuspended AMPure XP Beads (AXP, or SPRI) and mix by pipetting.
Example volume X24 samples X48 samples X96 samples Volume of 1X AXP 120 µl 240 µl 480 µl -
Incubate on a Hula mixer (rotator mixer) for 5 minutes at room temperature.
-
Prepare at least 3 ml of fresh 80% ethanol in nuclease-free water.
-
Spin down the sample and pellet on a magnet. Keep the tube on the magnet, and pipette off the supernatant when clear and colourless.
-
Keep the tube on the magnet and wash the beads with 1 ml of freshly-prepared 80% ethanol without disturbing the pellet. Remove the ethanol using a pipette and discard.
-
Repeat the previous step.
-
Briefly spin down and place the tube back on the magnet. Pipette off any residual ethanol. Allow to dry for 30 seconds, but do not dry the pellet to the point of cracking.
-
Remove the tube from the magnetic rack and resuspend the pellet by pipetting in 15 µl Elution Buffer (EB). Incubate for 10 minutes at room temperature.
-
Pellet the beads on a magnet until the eluate is clear and colourless.
-
Remove and retain 15 µl of eluate containing the DNA library into a clean 1.5 ml Eppendorf DNA LoBind tube.
-
Quantify DNA concentration by using the Qubit dsDNA HS Assay Kit.
-
Take forward 11 µl of your eluted DNA library.
-
In a fresh 1.5 ml Eppendorf DNA LoBind tube, dilute the Rapid Adapter (RA) as follows and pipette mix:
Reagent Volume Rapid Adapter (RA) 1.5 μl Adapter Buffer (ADB) 3.5 μl Total 5 μl -
Add 1 µl of the diluted Rapid Adapter (RA) to the barcoded DNA.
-
Mix gently by flicking the tubes, and spin down.
-
Incubate the reaction for 5 minutes at room temperature.
Priming and loading the SpotON flow cell
Priming and loading the SpotON flow cell
- Materials
-
- Flow Cell Flush (FCF)
- Flow Cell Tether (FCT)
- Library Solution (LIS)
- Library Beads (LIB)
- Sequencing Buffer (SB)
- Consumables
-
- 1.5 ml Eppendorf DNA LoBind tubes
- MinION and GridION Flow Cell
- Nuclease-free water (e.g. ThermoFisher, AM9937)
- Bovine Serum Albumin (BSA) (50 mg/ml) (e.g Invitrogen™ UltraPure™ BSA 50 mg/ml, AM2616)
- Equipment
-
- MinION or GridION device
- MinION and GridION Flow Cell Light Shield
- P1000 pipette and tips
- P100 pipette and tips
- P20 pipette and tips
- P10 pipette and tips
-
Using the Library Solution
For most sequencing experiments, use the Library Beads (LIB) for loading your library onto the flow cell. However, for viscous libraries it may be difficult to load with the beads and may be appropriate to load using the Library Solution (LIS).
-
Thaw the Sequencing Buffer (SB), Library Beads (LIB) or Library Solution (LIS, if using), Flow Cell Tether (FCT) and Flow Cell Flush (FCF) at room temperature before mixing by vortexing. Then spin down and store on ice.
-
Prepare the flow cell priming mix with BSA in a suitable tube for the number of flow cells to flush. Once combined, mix well by pipette mixing.
Reagents Volume per flow cell Flow Cell Flush (FCF) 1,170 µl Bovine Serum Albumin (BSA) at 50 mg/ml 5 µl Flow Cell Tether (FCT) 30 µl Total volume 1,205 µl -
Open the MinION or GridION device lid and slide the flow cell under the clip. Press down firmly on the priming port cover to ensure correct thermal and electrical contact.
-
Optional ActionComplete a flow cell check to assess the number of pores available before loading the library.
This step can be omitted if the flow cell has been checked previously.
See the flow cell check instructions in the MinKNOW protocol for more information.
-
Slide the flow cell priming port cover clockwise to open the priming port.
-
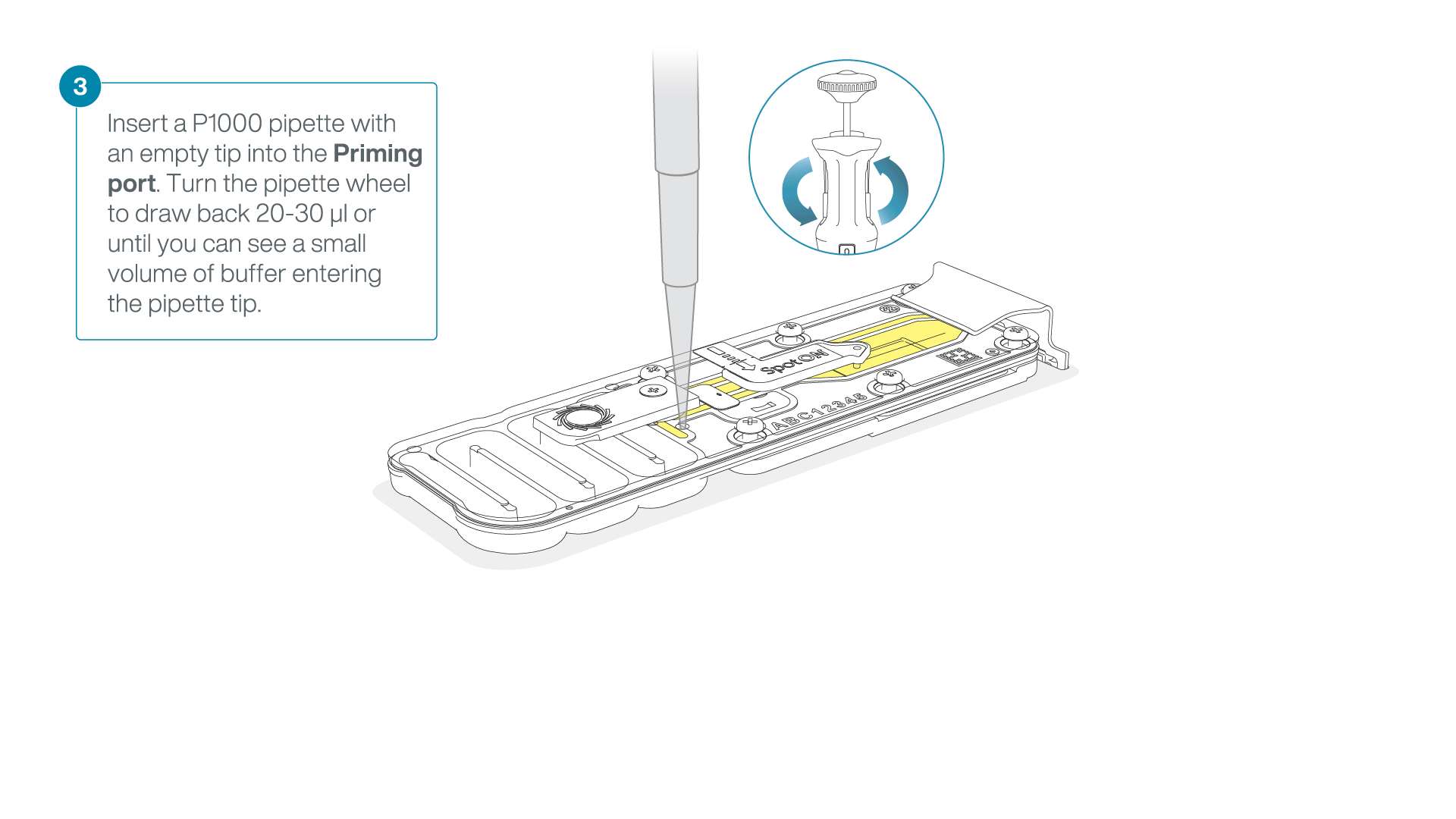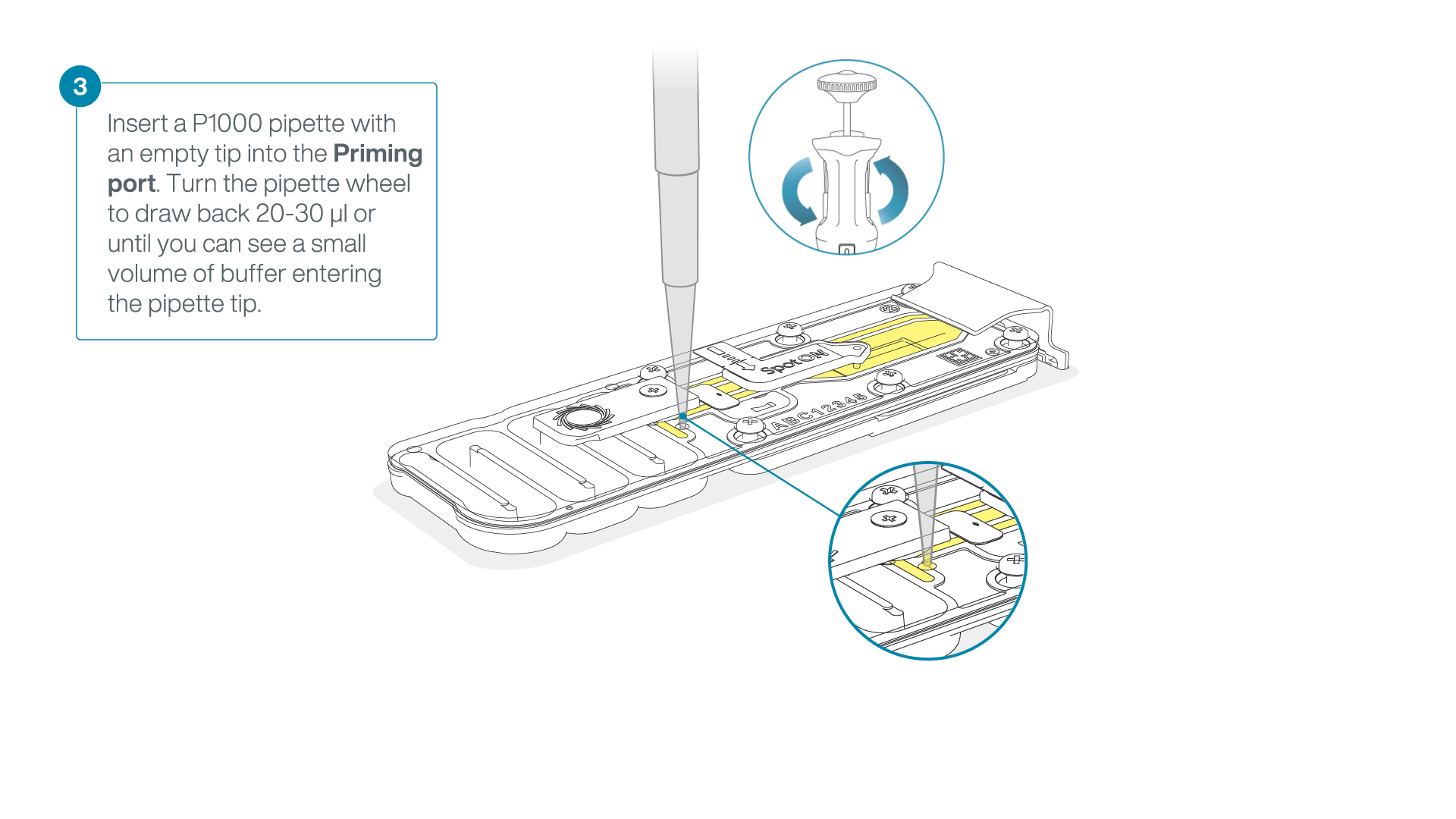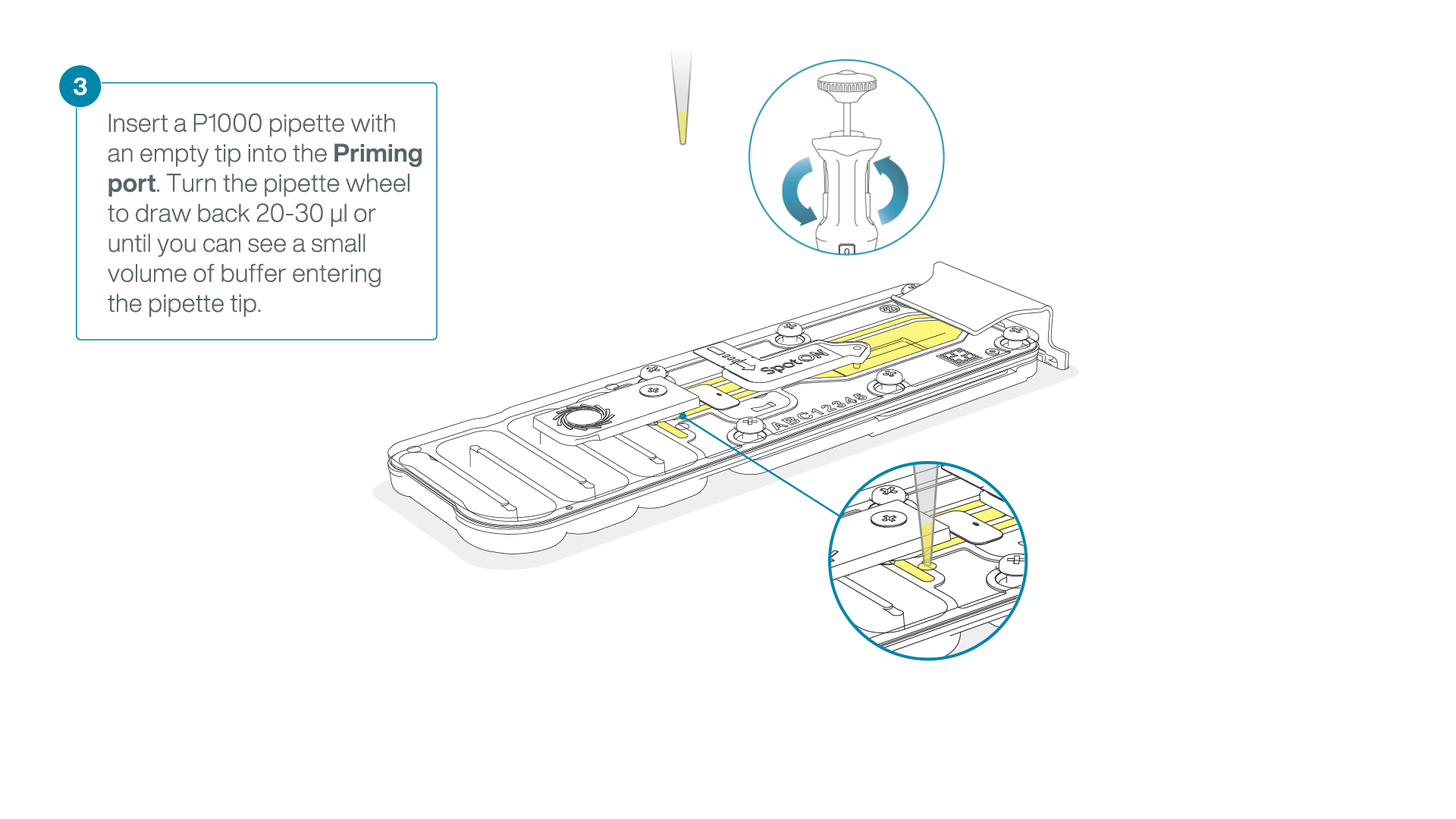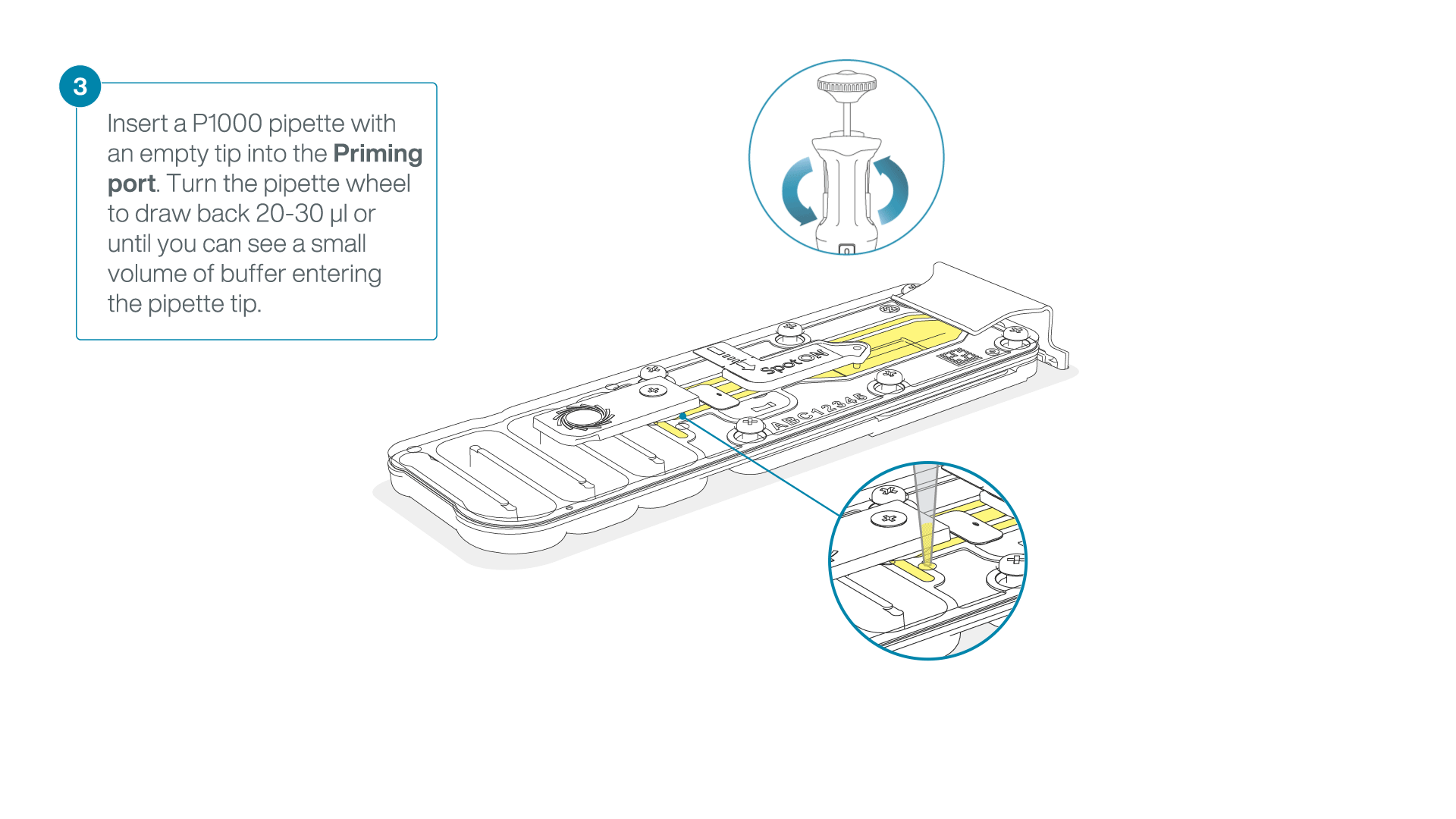
After opening the priming port, check for a small air bubble under the cover. Draw back a small volume to remove any bubbles:
- Set a P1000 pipette to 200 µl
- Insert the tip into the priming port
- Turn the wheel until the dial shows 220-230 µl, to draw back 20-30 µl, or until you can see a small volume of buffer entering the pipette tip
Note: Visually check that there is continuous buffer from the priming port across the sensor array.
-
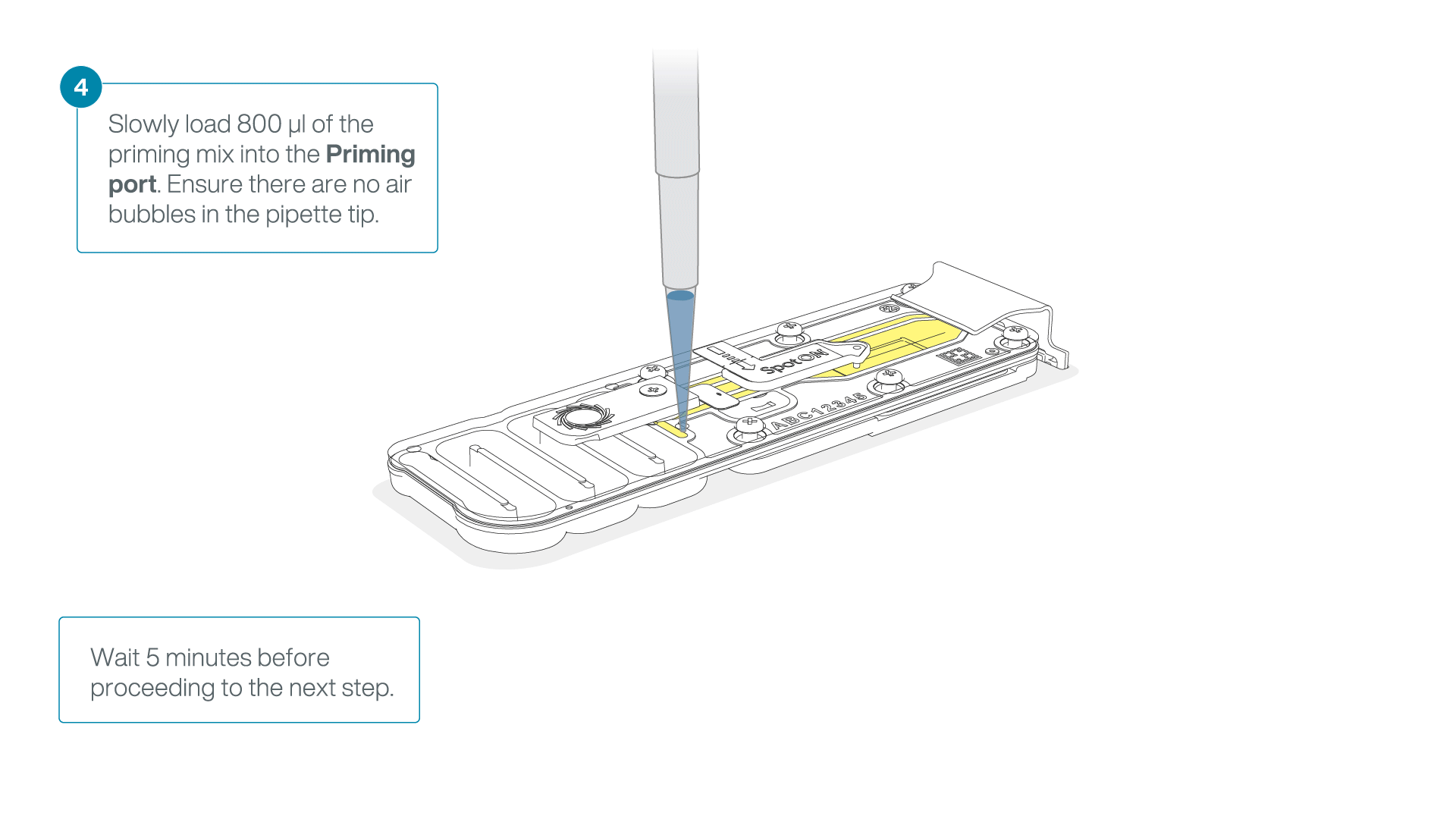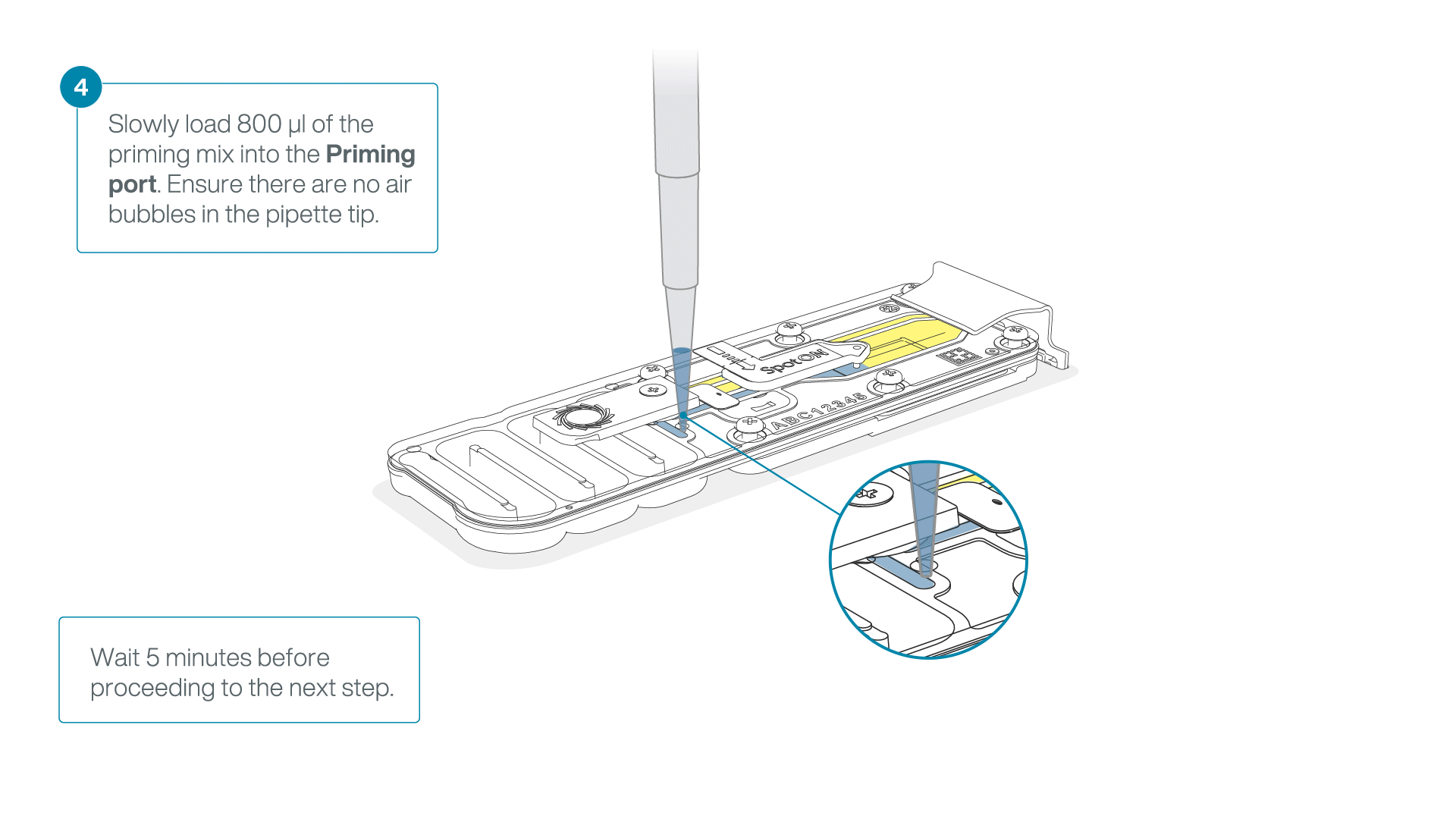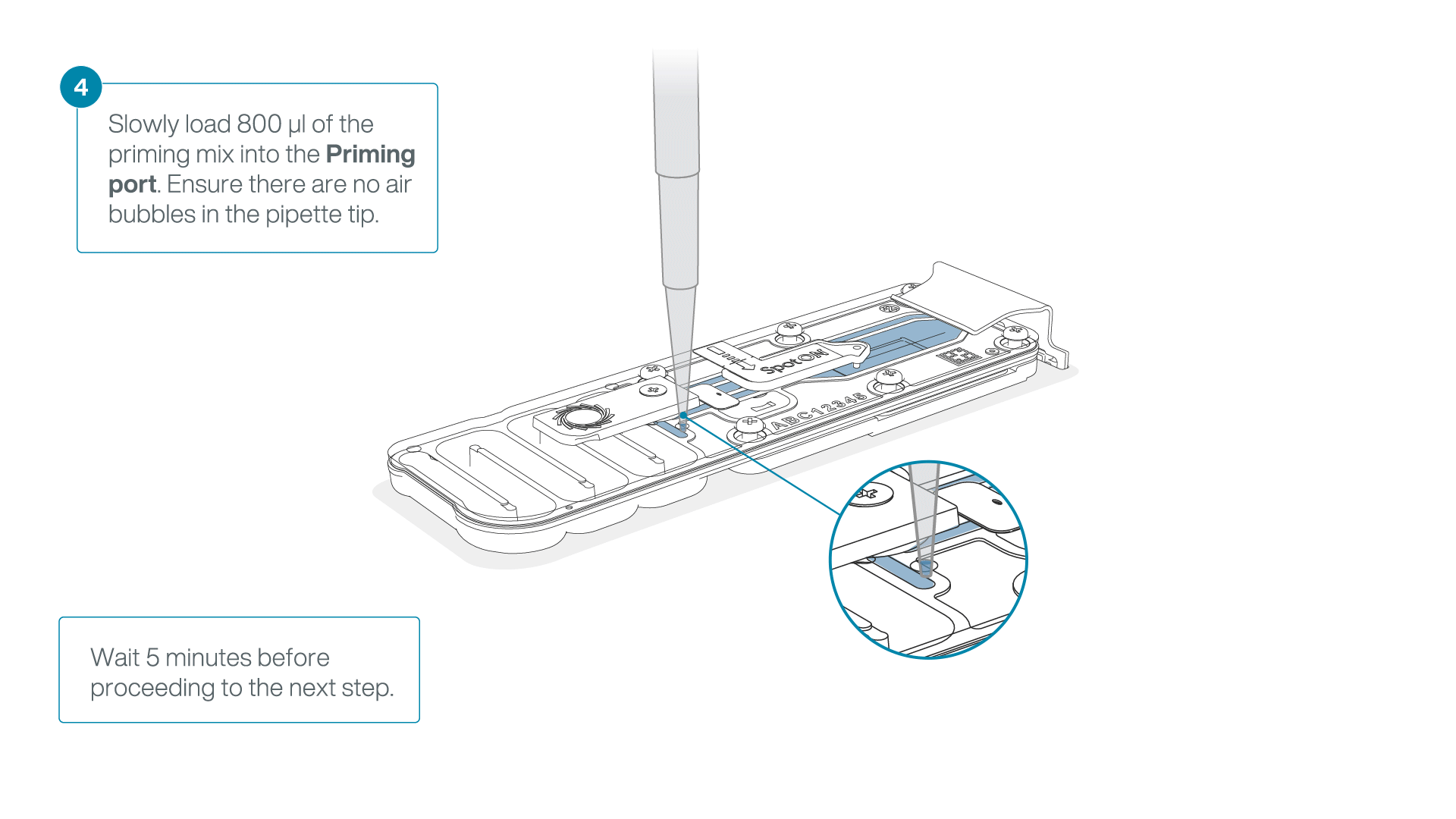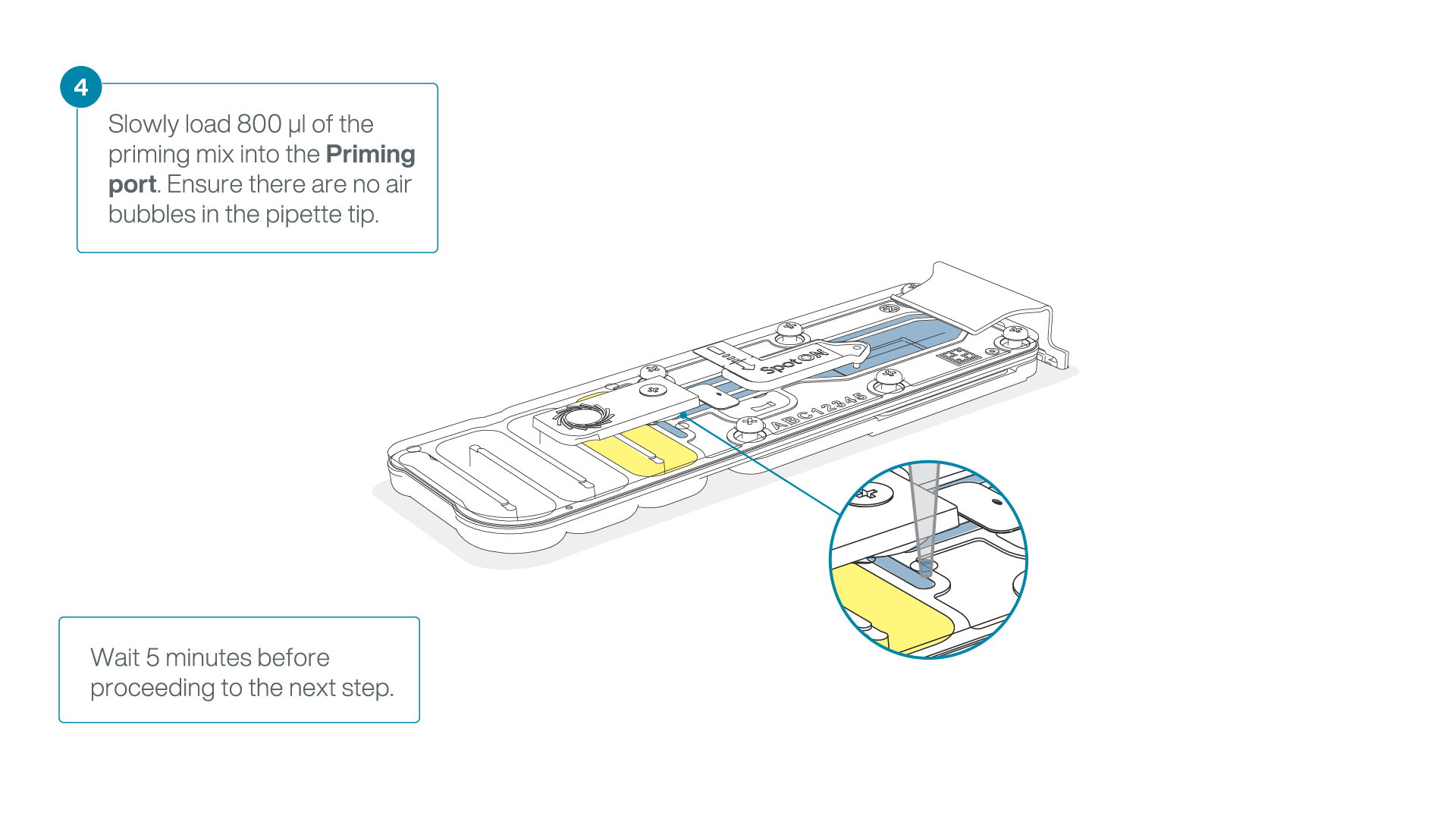
Load 800 µl of the priming mix into the flow cell via the priming port, avoiding the introduction of air bubbles. Wait for five minutes. During this time, prepare the library for loading by following the steps below.
-
Thoroughly mix the contents of the Library Beads (LIB) by pipetting.
-
In a new 1.5 ml Eppendorf DNA LoBind tube, prepare the library for loading as follows:
Reagent Volume per flow cell Sequencing Buffer (SB) 37.5 µl Library Beads (LIB) mixed immediately before use, or Library Solution (LIS), if using 25.5 µl DNA library 12 µl Total 75 µl -
Complete the flow cell priming:
- Gently lift the SpotON sample port cover to make the SpotON sample port accessible.
- Load 200 µl of the priming mix into the flow cell priming port (not the SpotON sample port), avoiding the introduction of air bubbles.
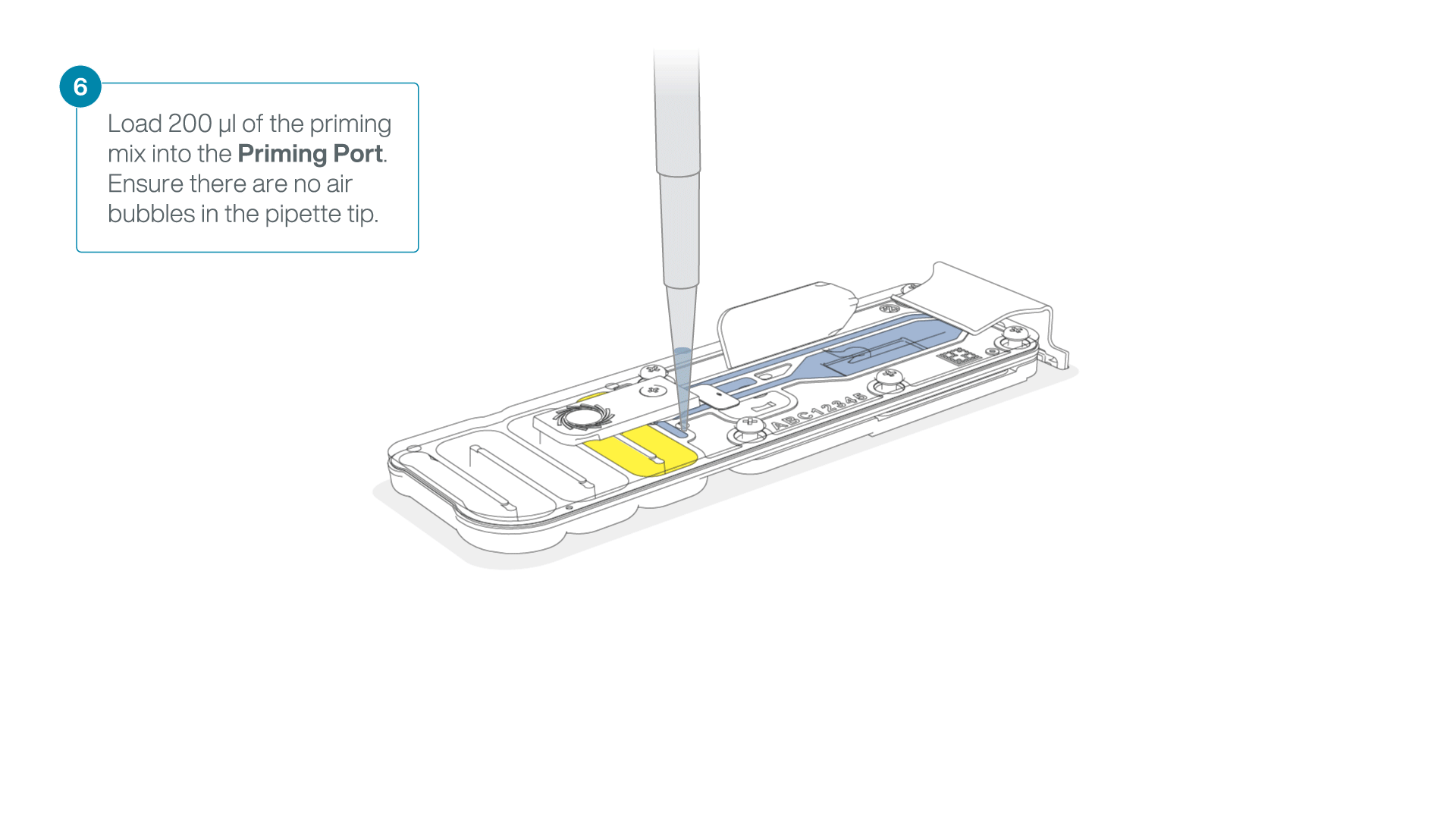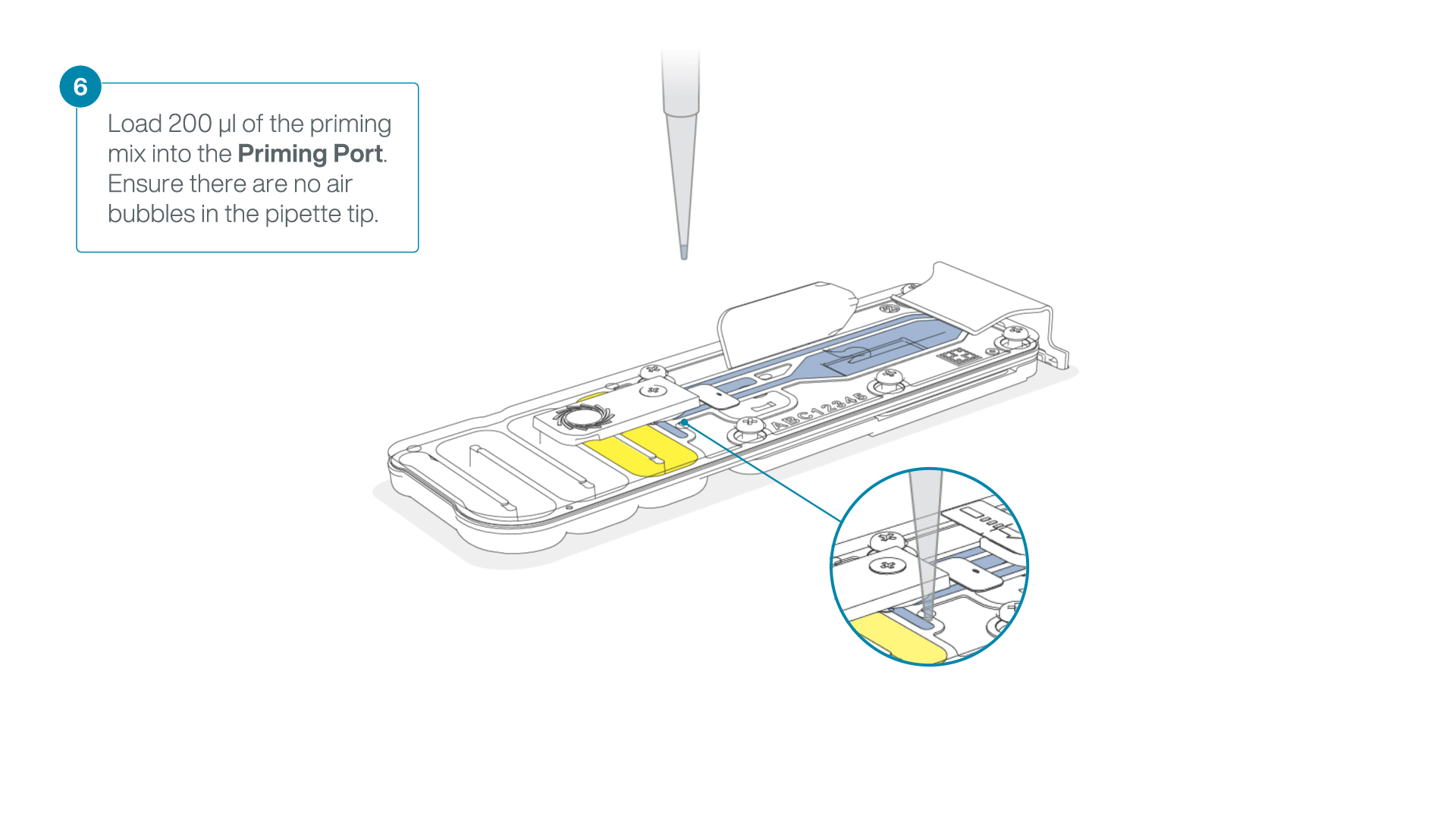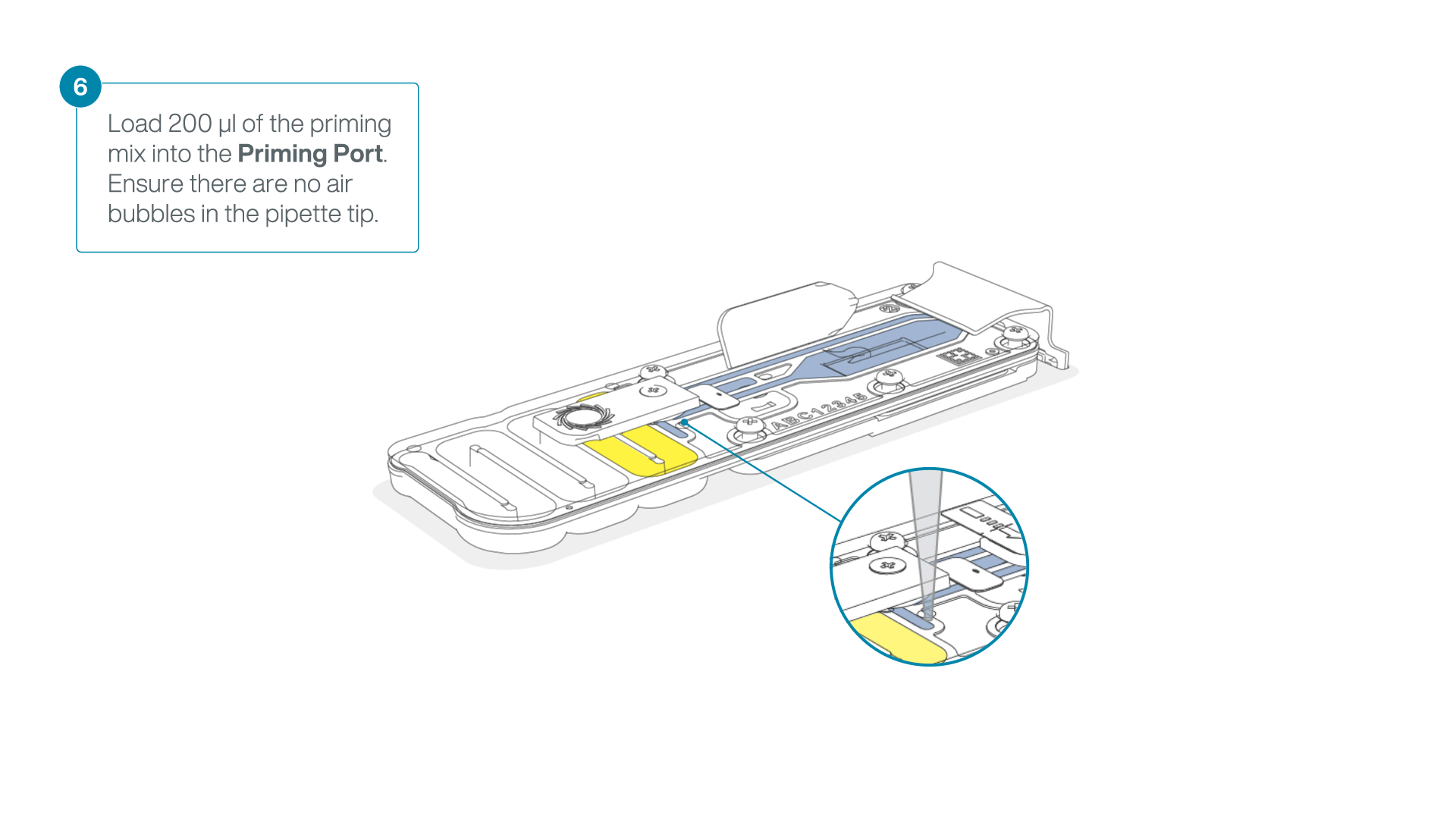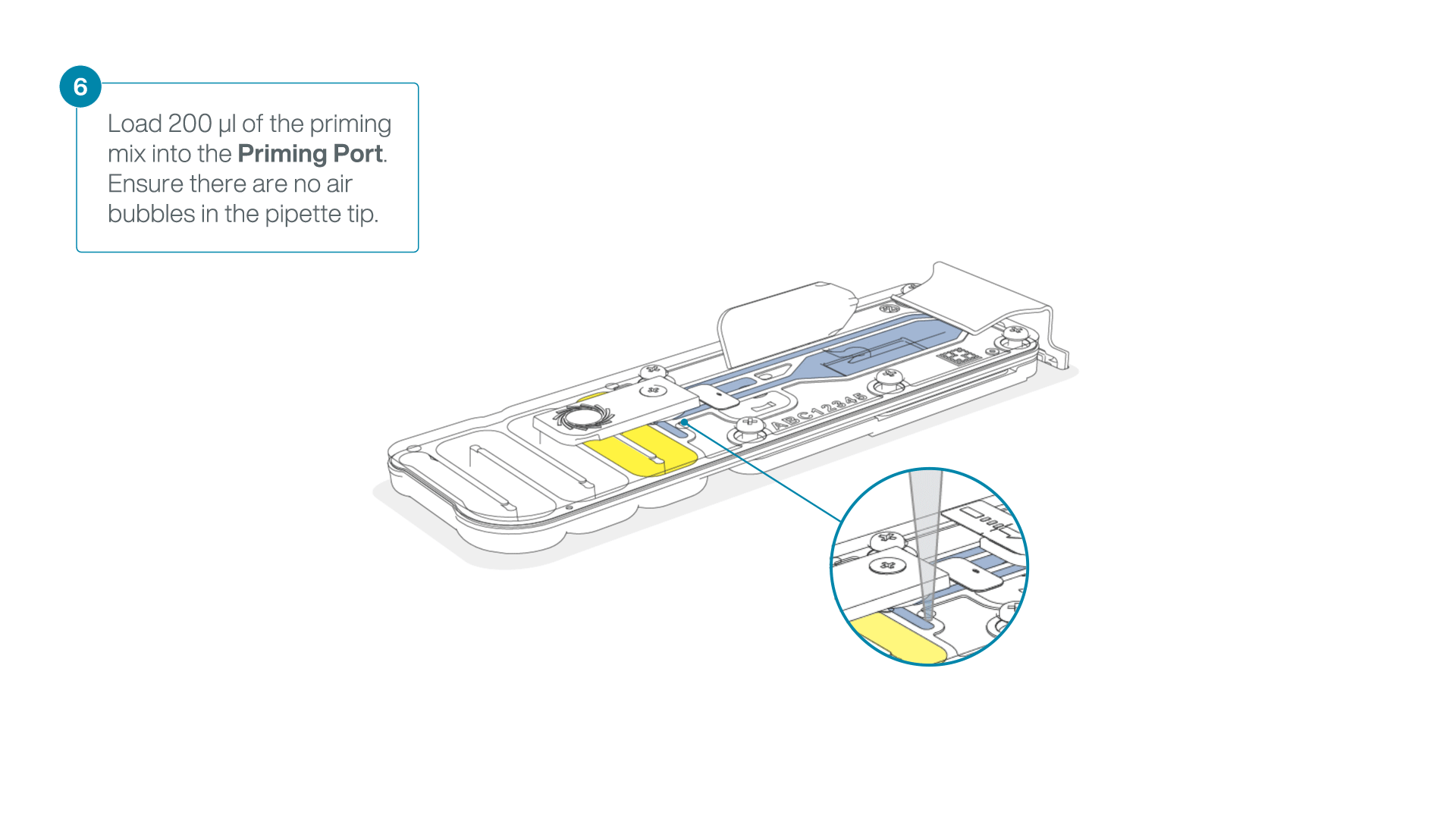
-
Mix the prepared library gently by pipetting up and down just prior to loading.
-
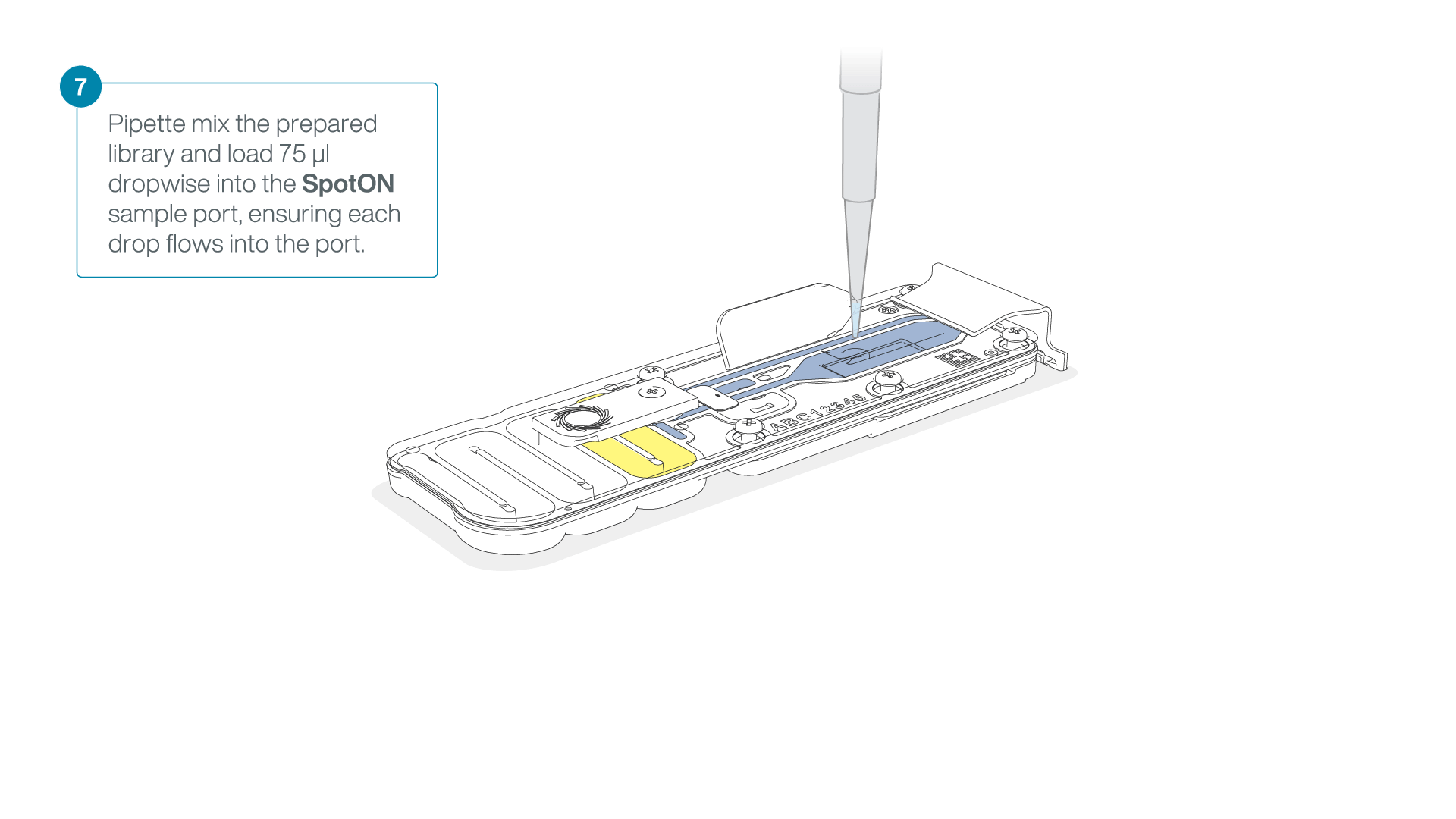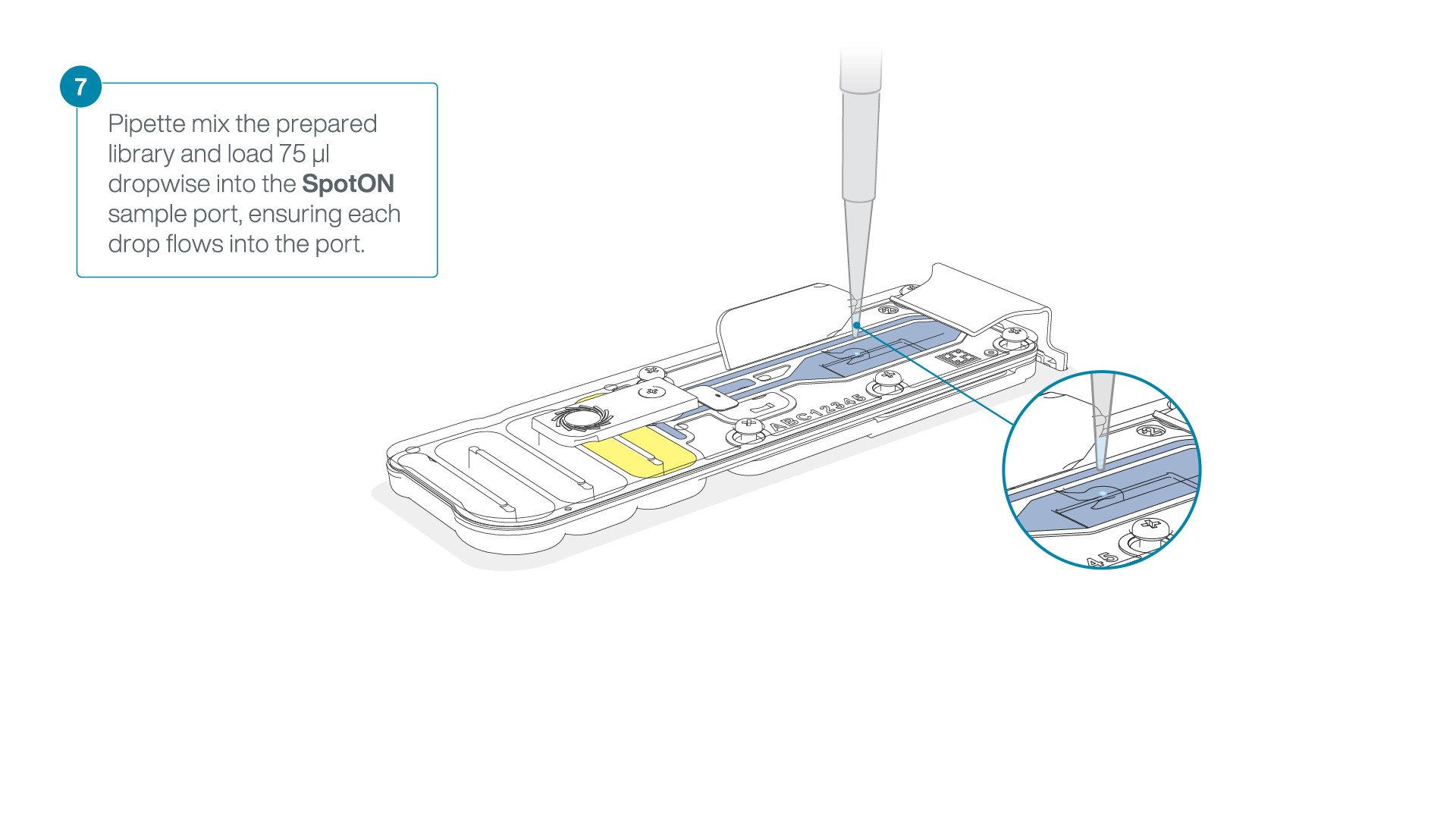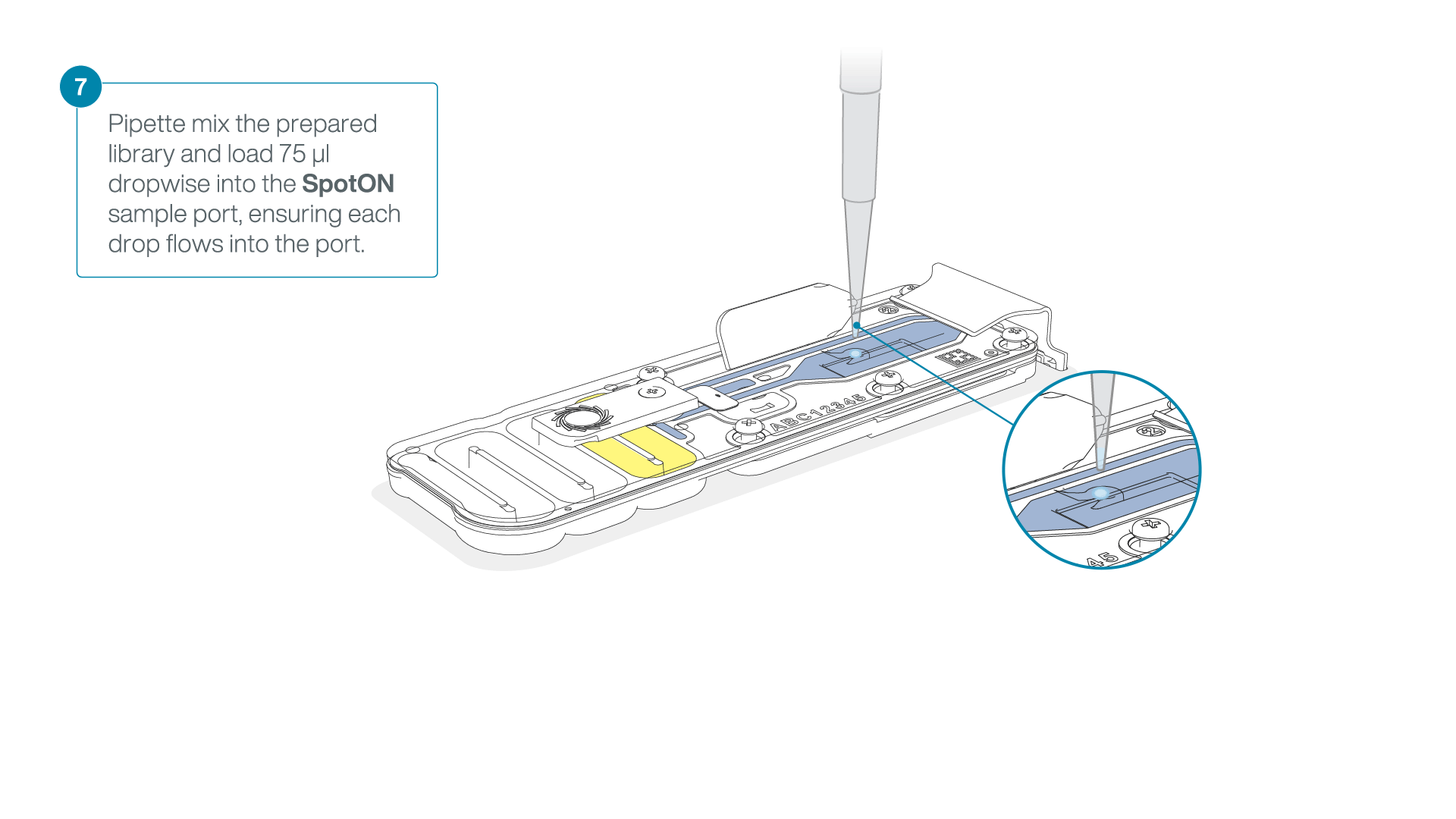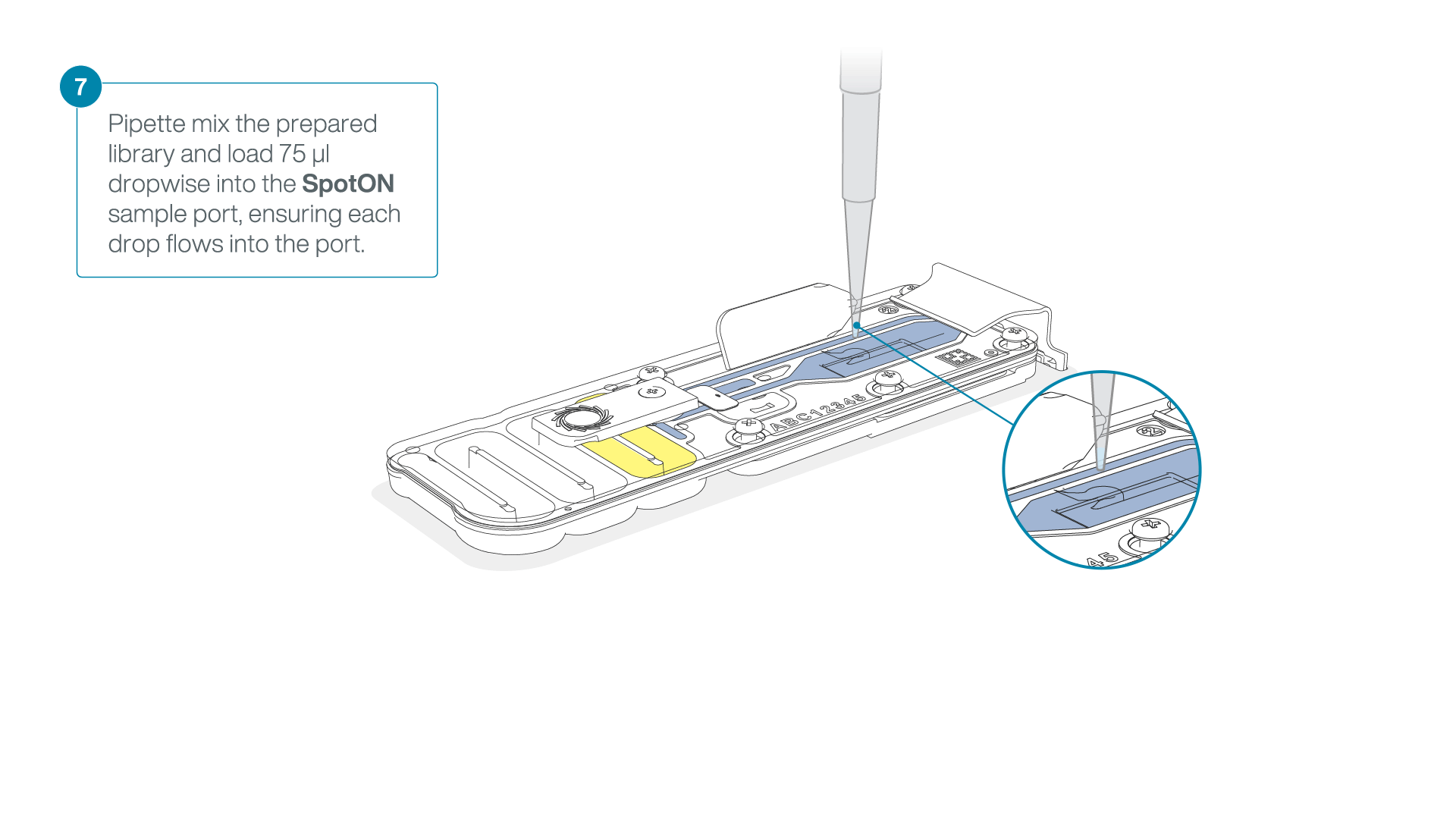
Add 75 μl of the prepared library to the flow cell via the SpotON sample port in a dropwise fashion. Ensure each drop flows into the port before adding the next.
-
Gently replace the SpotON sample port cover, making sure the bung enters the SpotON port and close the priming port.

-
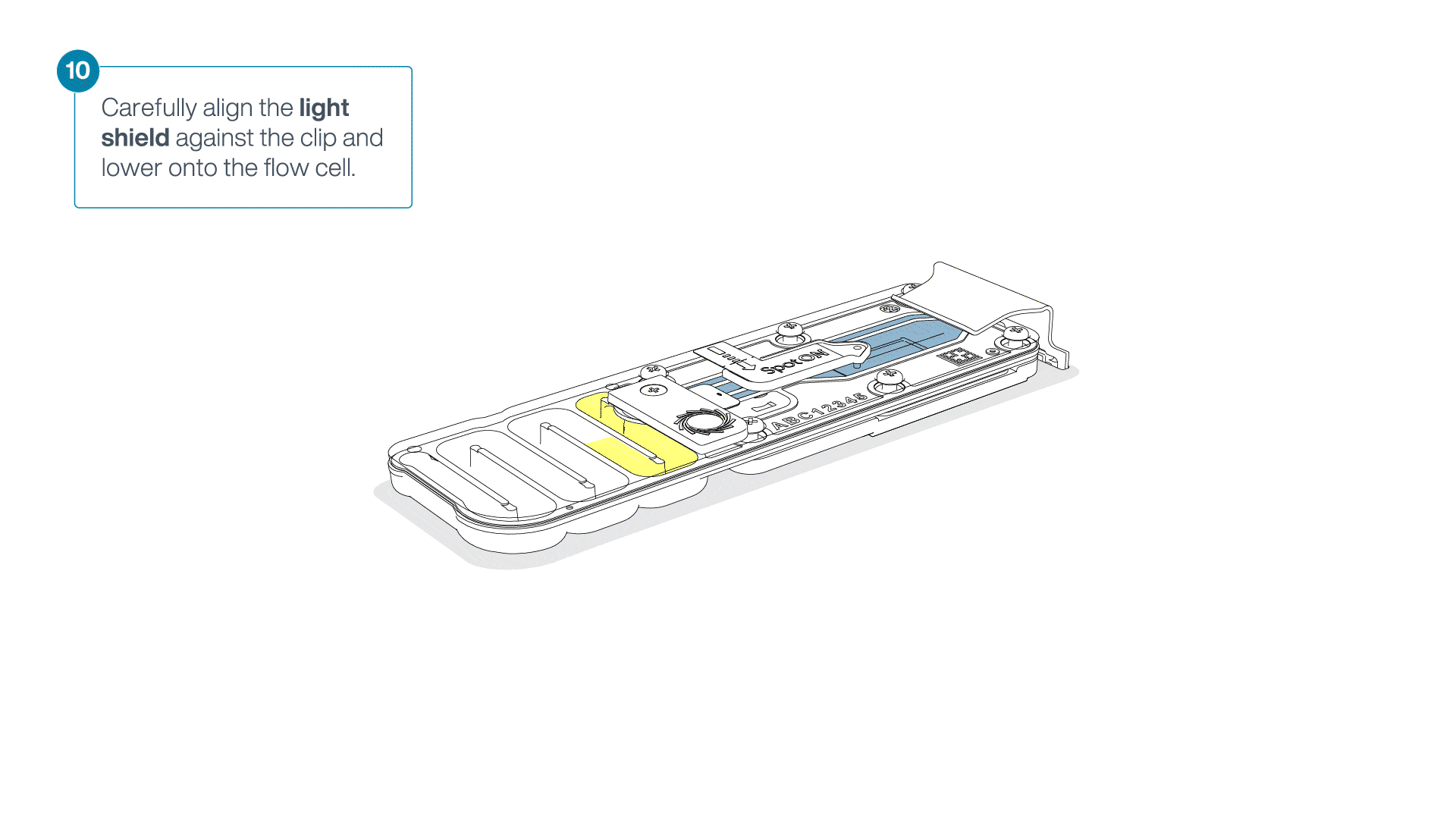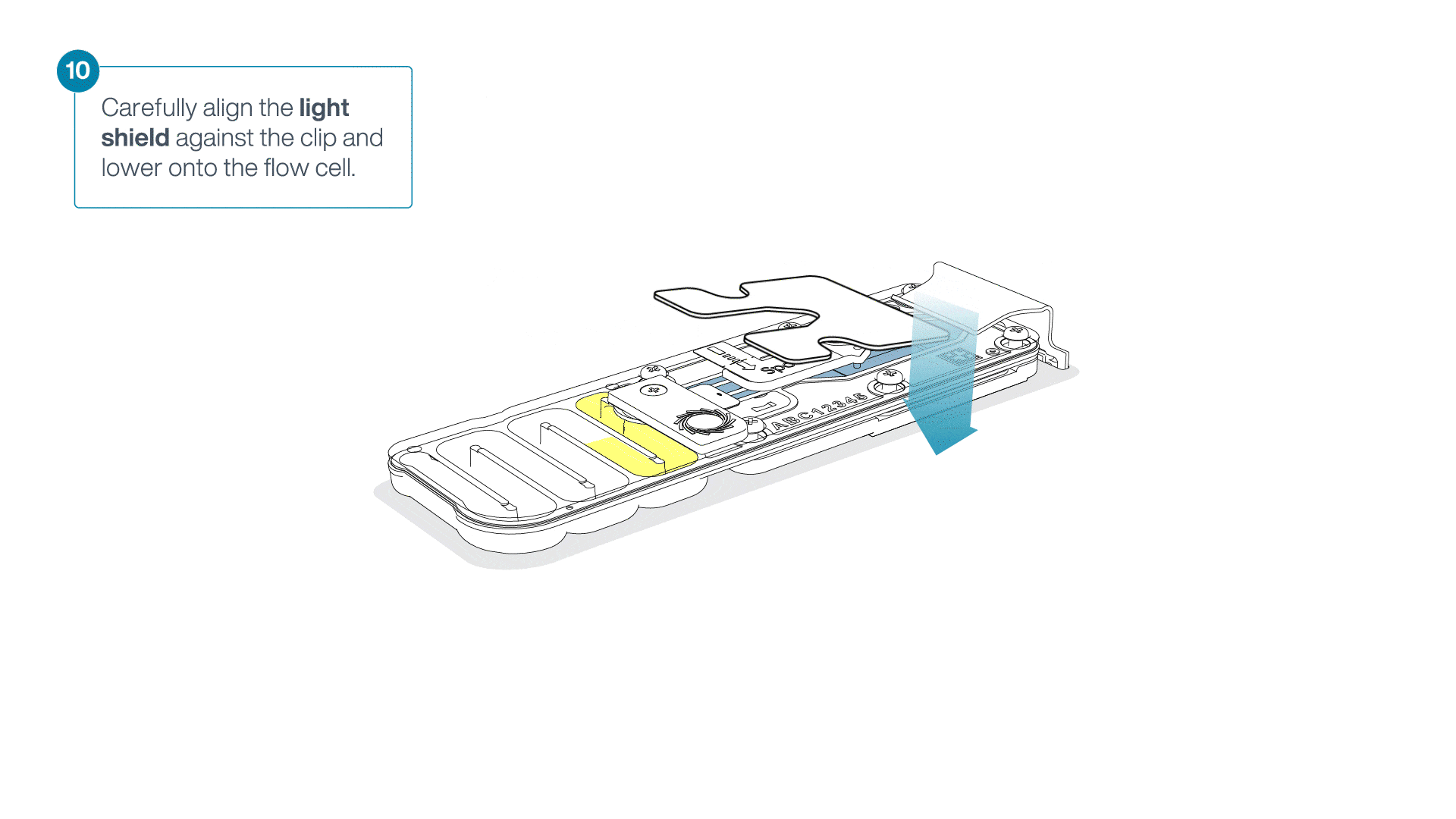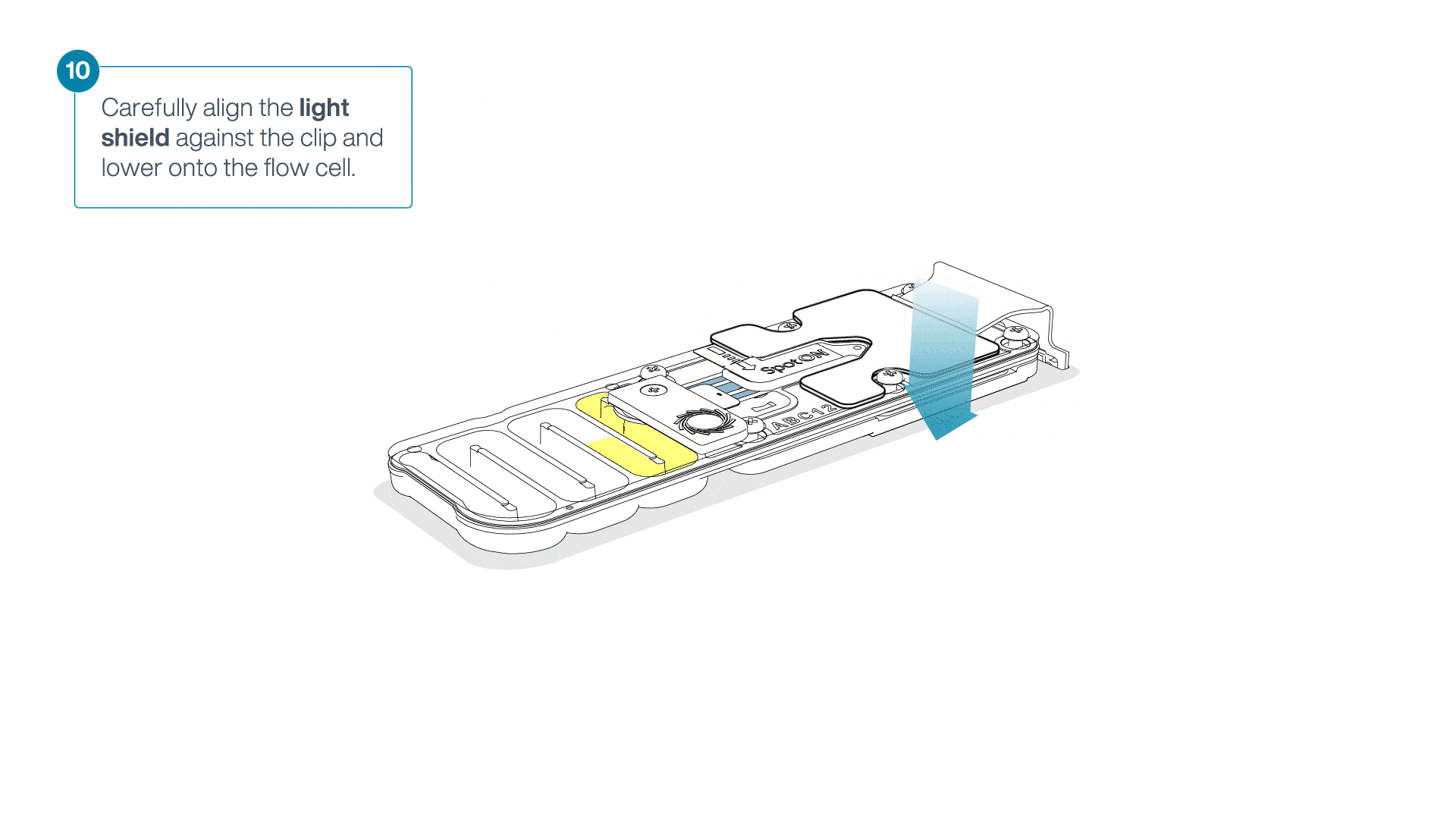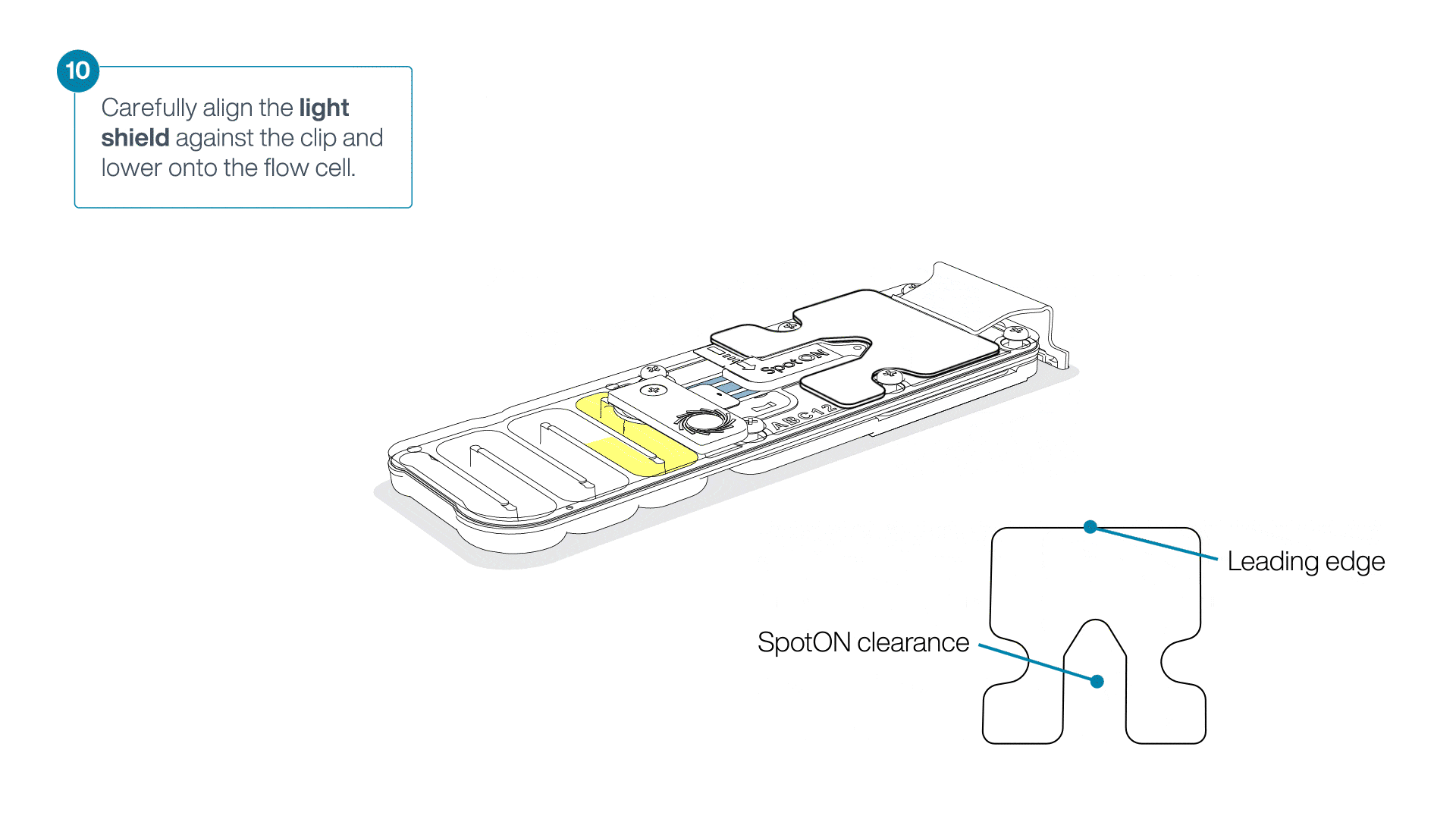
Place the light shield onto the flow cell, as follows:
Carefully place the leading edge of the light shield against the clip.
Note: Do not force the light shield underneath the clip.Gently lower the light shield onto the flow cell. The light shield should sit around the SpotON cover, covering the entire top section of the flow cell.
Sequencing and data analysis
Data acquisition and basecalling
Data acquisition and basecalling
-
Overview of nanopore data analysis
For a full overview of nanopore data analysis, which includes options for basecalling and post-basecalling analysis, please refer to the Data Analysis document.
-
How to start sequencing
The sequencing device control, data acquisition and real-time basecalling are carried out by the MinKNOW software. Please ensure MinKNOW is installed on your computer or device. There are multiple options for how to carry out sequencing:
1. Data acquisition and basecalling in real-time using MinKNOW on a computer
Follow the instructions in the MinKNOW protocol beginning from the "Starting a sequencing run" section until the end of the "Completing a MinKNOW run" section.
2. Data acquisition and basecalling in real-time using the MinION Mk1B/Mk1D device
Follow the instructions in the MinION Mk1B user manual or the MinION Mk1D user manual.
3. Data acquisition and basecalling in real-time using the MinION Mk1C device
Follow the instructions in the MinION Mk1C user manual.
4. Data acquisition and basecalling in real-time using the GridION device
Follow the instructions in the GridION user manual.
5. Data acquisition and basecalling in real-time using the PromethION device
Follow the instructions in the PromethION user manual or the PromethION 2 Solo user manual.
6. Data acquisition using MinKNOW on a computer and basecalling at a later time using MinKNOW
Follow the instructions in the MinKNOW protocol beginning from the "Starting a sequencing run" section until the end of the "Completing a MinKNOW run" section. When setting your experiment parameters, set the Basecalling tab to OFF. After the sequencing experiment has completed, follow the instructions in the Post-run analysis section of the MinKNOW protocol.
Downstream analysis
Downstream analysis
-
Recommended pipeline analysis
The wf-artic is a bioinformatics workflow for the analysis of ARTIC sequencing data prepared using the Midnight protocol. The bioinformatics workflow is orchestrated by the Nextflow software. Nextflow is a publicly available and open-source project that enables the execution of scientific workflows in a scalable and reproducible way. The use of the Nextflow software has been integrated into the EPI2ME Labs software that we recommend for running our downstream analysis methods.
Alternative methods for downstream analysis are available using your device terminal or command line, however we only suggest this for experienced users.
Demultiplexed sequence reads are processed using the ARTIC Field Bioinformatics software that has been modified for the analysis of FASTQ sequences prepared using Oxford Nanopore Rapid Sequencing kits. The other modification to the ARTIC workflow is the use of a primer scheme that defines the sequencing primers used by the Midnight protocol and their genomic locations on the SARS-CoV-2 genome.
The wf-artic workflow includes other analytical steps that include cladistic analysis using Nextclade and strain assignment using Pangolin. The data facets included in the report are parameterised and additional information such as plots of depth-of-coverage across the reference genome is optional.
The complete source for wf-artic is linked, and the Nextflow software will download the scripts and logic flow from this location.
The wf-artic workflow needs to be started manually as outlined below in 'Running a Midnight analysis using EPI2ME Labs'.
-
Software set-up and installation
The EPI2ME application provides a clean interface to accessing bioinformatics workflows, and is our recommended method in performing your post-sequencing analysis.
Follow the instructions in the EPI2ME Installation guide to install the application on your device.
For more information on how to use EPI2ME, refer to the EPI2ME Quick Start guide.
-
Installing and updating the wf-artic workflow in EPI2ME Labs:
Ensure you have installed the wf-artic workflow prior to the first analysis set-up.
In the EPI2ME Labs home page, scroll down to the "Install workflows" section and click on epi2me-labs/wf-artic:
If you have already installed the wf-artic workflow, ensure you are using the latest version.
Updating the workflow can be done directly through EPI2ME Labs by navigating to the wf-artic workflow page and clicking Update Workflow:
-
Demultiplexing of multiple barcoded samples
The wf-artic analysis requires FASTQ sequence data that has already been demultiplexed.
Reads will be demultiplexed during sequencing if you are following the recommended "Required settings in MinKNOW". However, demultiplexing can also be done post-sequencing using the MinKNOW software.
For more information and guides on demultiplexing using MinKNOW, refer to the "Post-run analysis" section in our MinKNOW Protocol.
The expected input for wf-artic is a folder of folders as shown below. Each of the barcode folders should contain the FASTQ sequence data and files may either be uncompressed or gzipped.
$ tree -d MidnightFastq/MidnightFastq/├── barcode01├── barcode02├── barcode03├── barcode04├── barcode05├── barcode06└── unclassified -
Running a Midnight analysis using EPI2ME Labs
-
Open the EPI2ME Labs application on your device.
-
Open the "Workflows" tab in the EPI2ME Labs application and click on the "wf-artic" workflow:
-
In the "wf-artic" workflow page, select "Run this workflow" to open analysis set-up:
-
Complete the wf-artic run set-up:
Select your data input file location. Please note, this folder must contain the demultiplexed FASTQ files of your sequencing run.
Expand the Primer Scheme Selection tab and set the Scheme version to Midnight-ONT/V3.
Expand the Advanced Options tab and set the Medaka model to the basecalling model used in your sequencing run.
Expand the Extra configuration tab and set the Run name for your wf-artic analysis.
Click Launch workflow at the bottom of the page to begin your analysis.
-
Completed analysis and result files
The wf-artic analysis outputs will be written to the Working Directory folder specified in the EPI2ME Labs Settings tab.
The location of this folder is specified in the wf-artic run Instance parameters preceeded byout_dir.However, these files can also be accessed directly in the EPI2ME Labs application from the completed analysis page for your run:
These outputs include:
all_consensus.fasta
A multi-FASTA format sequence file containing the consensus sequence for each of the samples investigated. This consensus sequence has been prepared for the whole SARS-CoV-2 genome, not just the spike protein region. The consensus sequence masks the non-spike regions and regions of low sequence coverage with N residues.all_variants.vcf.gz
A gzipped VCF file that describes all high-quality genetic variants called by medaka from the sequenced samples.all_variants.vcf.gz.tbi
An index file for the gzipped VCF file.consensus_status.txt
A tab delimited file that reports whether a consensus sequence has been successfully prepared for a sample, or not.wf-artic-report.html
A report summarising these data. This HTML format report also includes the output of the Nextclade software that can be used for a visual inspection of, for example, primer drop out or other qualitative consensus sequence aspects.
Other files are included in the
work-directory. This includes per sample VCF files of all genetic variants prior to filtering and other sequences. -
Housekeeping and disk usage
The "Working Directory" can be specified in the EPI2ME Labs "Settings" tab and defines where the workflow intermediate files and outputs are stored.
This folder will accumulate a significant number of files that correspond to raw BAM files, other larger intermediates and analysis results files. We recommend this folder to be routinely cleared.
Flow cell reuse and returns
Flow cell reuse and returns
- Materials
-
- Flow Cell Wash Kit (EXP-WSH004)
-
After your sequencing experiment is complete, if you would like to reuse the flow cell, please follow the Flow Cell Wash Kit protocol and store the washed flow cell at +2°C to +8°C.
The Flow Cell Wash Kit protocol is available on the Nanopore Community.
-
Alternatively, follow the returns procedure to send the flow cell back to Oxford Nanopore.
Instructions for returning flow cells can be found here.
Troubleshooting
Issues during DNA/RNA extraction and library preparation
Issues during DNA/RNA extraction and library preparation
-
Below is a list of the most commonly encountered issues, with some suggested causes and solutions.
We also have an FAQ section available on the Nanopore Community Support section.
If you have tried our suggested solutions and the issue still persists, please contact Technical Support via email (support@nanoporetech.com) or via LiveChat in the Nanopore Community.
-
Low sample quality
Observation Possible cause Comments and actions Low DNA purity (Nanodrop reading for DNA OD 260/280 is <1.8 and OD 260/230 is <2.0–2.2) The DNA extraction method does not provide the required purity The effects of contaminants are shown in the Contaminants document. Please try an alternative extraction method that does not result in contaminant carryover.
Consider performing an additional SPRI clean-up step.Low RNA integrity (RNA integrity number <9.5 RIN, or the rRNA band is shown as a smear on the gel) The RNA degraded during extraction Try a different RNA extraction method. For more info on RIN, please see the RNA Integrity Number document. Further information can be found in the DNA/RNA Handling page. RNA has a shorter than expected fragment length The RNA degraded during extraction Try a different RNA extraction method. For more info on RIN, please see the RNA Integrity Number document. Further information can be found in the DNA/RNA Handling page.
We recommend working in an RNase-free environment, and to keep your lab equipment RNase-free when working with RNA. -
Low DNA recovery after AMPure bead clean-up
Observation Possible cause Comments and actions Low recovery DNA loss due to a lower than intended AMPure beads-to-sample ratio 1. AMPure beads settle quickly, so ensure they are well resuspended before adding them to the sample.
2. When the AMPure beads-to-sample ratio is lower than 0.4:1, DNA fragments of any size will be lost during the clean-up.Low recovery DNA fragments are shorter than expected The lower the AMPure beads-to-sample ratio, the more stringent the selection against short fragments. Please always determine the input DNA length on an agarose gel (or other gel electrophoresis methods) and then calculate the appropriate amount of AMPure beads to use. 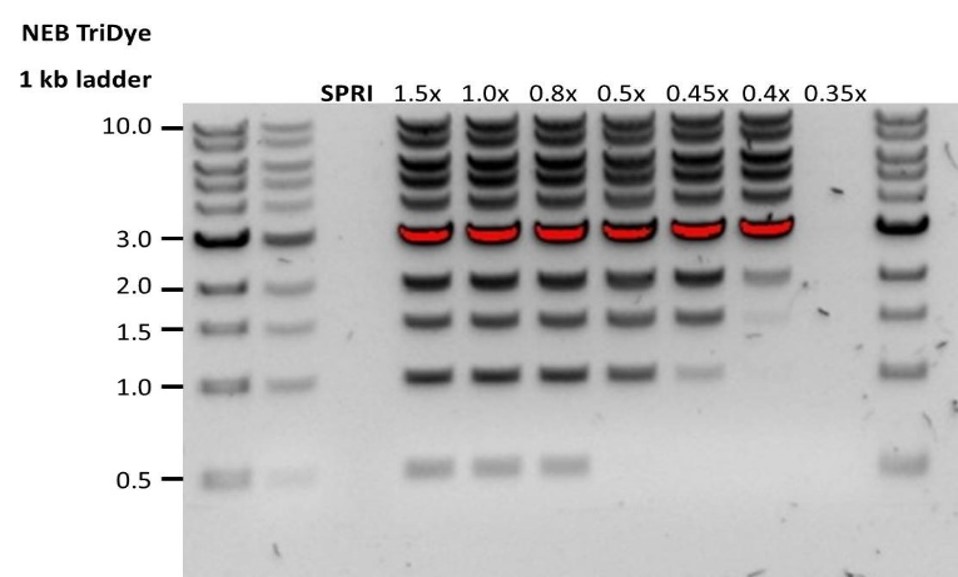
Low recovery after end-prep The wash step used ethanol <70% DNA will be eluted from the beads when using ethanol <70%. Make sure to use the correct percentage.
Issues during the sequencing run
Issues during the sequencing run
-
Below is a list of the most commonly encountered issues, with some suggested causes and solutions.
We also have an FAQ section available on the Nanopore Community Support section.
If you have tried our suggested solutions and the issue still persists, please contact Technical Support via email (support@nanoporetech.com) or via LiveChat in the Nanopore Community.
-
Fewer pores at the start of sequencing than after Flow Cell Check
Observation Possible cause Comments and actions MinKNOW reported a lower number of pores at the start of sequencing than the number reported by the Flow Cell Check An air bubble was introduced into the nanopore array After the Flow Cell Check it is essential to remove any air bubbles near the priming port before priming the flow cell. If not removed, the air bubble can travel to the nanopore array and irreversibly damage the nanopores that have been exposed to air. The best practice to prevent this from happening is demonstrated in this video. MinKNOW reported a lower number of pores at the start of sequencing than the number reported by the Flow Cell Check The flow cell is not correctly inserted into the device Stop the sequencing run, remove the flow cell from the sequencing device and insert it again, checking that the flow cell is firmly seated in the device and that it has reached the target temperature. If applicable, try a different position on the device (GridION/PromethION). MinKNOW reported a lower number of pores at the start of sequencing than the number reported by the Flow Cell Check Contaminations in the library damaged or blocked the pores The pore count during the Flow Cell Check is performed using the QC DNA molecules present in the flow cell storage buffer. At the start of sequencing, the library itself is used to estimate the number of active pores. Because of this, variability of about 10% in the number of pores is expected. A significantly lower pore count reported at the start of sequencing can be due to contaminants in the library that have damaged the membranes or blocked the pores. Alternative DNA/RNA extraction or purification methods may be needed to improve the purity of the input material. The effects of contaminants are shown in the Contaminants Know-how piece. Please try an alternative extraction method that does not result in contaminant carryover. -
MinKNOW script failed
Observation Possible cause Comments and actions MinKNOW shows "Script failed" Restart the computer and then restart MinKNOW. If the issue persists, please collect the MinKNOW log files and contact Technical Support. If you do not have another sequencing device available, we recommend storing the flow cell and the loaded library at 4°C and contact Technical Support for further storage guidance. -
Pore occupancy below 40%
Observation Possible cause Comments and actions Pore occupancy <40% Not enough library was loaded on the flow cell Ensure you load the recommended amount of good quality library in the relevant library prep protocol onto your flow cell. Please quantify the library before loading and calculate mols using tools like the Promega Biomath Calculator, choosing "dsDNA: µg to pmol" Pore occupancy close to 0 The Ligation Sequencing Kit was used, and sequencing adapters did not ligate to the DNA Make sure to use the NEBNext Quick Ligation Module (E6056) and Oxford Nanopore Technologies Ligation Buffer (LNB, provided in the sequencing kit) at the sequencing adapter ligation step, and use the correct amount of each reagent. A Lambda control library can be prepared to test the integrity of the third-party reagents. Pore occupancy close to 0 The Ligation Sequencing Kit was used, and ethanol was used instead of LFB or SFB at the wash step after sequencing adapter ligation Ethanol can denature the motor protein on the sequencing adapters. Make sure the LFB or SFB buffer was used after ligation of sequencing adapters. Pore occupancy close to 0 No tether on the flow cell Tethers are adding during flow cell priming (FLT/FCT tube). Make sure FLT/FCT was added to FB/FCF before priming. -
Shorter than expected read length
Observation Possible cause Comments and actions Shorter than expected read length Unwanted fragmentation of DNA sample Read length reflects input DNA fragment length. Input DNA can be fragmented during extraction and library prep.
1. Please review the Extraction Methods in the Nanopore Community for best practice for extraction.
2. Visualise the input DNA fragment length distribution on an agarose gel before proceeding to the library prep.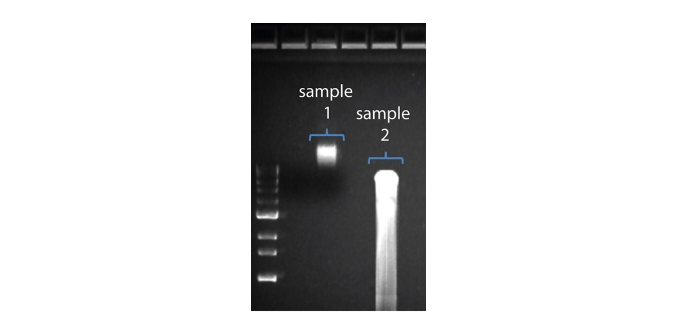 In the image above, Sample 1 is of high molecular weight, whereas Sample 2 has been fragmented.
In the image above, Sample 1 is of high molecular weight, whereas Sample 2 has been fragmented.
3. During library prep, avoid pipetting and vortexing when mixing reagents. Flicking or inverting the tube is sufficient. -
Large proportion of inactive pores
Observation Possible cause Comments and actions Large proportion of inactive/unavailable pores (shown as light blue in the channels panel and pore activity plot. Pores or membranes are irreversibly damaged) Air bubbles have been introduced into the flow cell Air bubbles introduced through flow cell priming and library loading can irreversibly damage the pores. Watch the Priming and loading your flow cell video for best practice Large proportion of inactive/unavailable pores Certain compounds co-purified with DNA Known compounds, include polysaccharides, typically associate with plant genomic DNA.
1. Please refer to the Plant leaf DNA extraction method.
2. Clean-up using the QIAGEN PowerClean Pro kit.
3. Perform a whole genome amplification with the original gDNA sample using the QIAGEN REPLI-g kit.Large proportion of inactive/unavailable pores Contaminants are present in the sample The effects of contaminants are shown in the Contaminants Know-how piece. Please try an alternative extraction method that does not result in contaminant carryover. -
Reduction in sequencing speed and q-score later into the run
Observation Possible cause Comments and actions Reduction in sequencing speed and q-score later into the run For Kit 9 chemistry (e.g. SQK-LSK109), fast fuel consumption is typically seen when the flow cell is overloaded with library (please see the appropriate protocol for your DNA library to see the recommendation). Add more fuel to the flow cell by following the instructions in the MinKNOW protocol. In future experiments, load lower amounts of library to the flow cell. -
Temperature fluctuation
Observation Possible cause Comments and actions Temperature fluctuation The flow cell has lost contact with the device Check that there is a heat pad covering the metal plate on the back of the flow cell. Re-insert the flow cell and press it down to make sure the connector pins are firmly in contact with the device. If the problem persists, please contact Technical Services. -
Failed to reach target temperature
Observation Possible cause Comments and actions MinKNOW shows "Failed to reach target temperature" The instrument was placed in a location that is colder than normal room temperature, or a location with poor ventilation (which leads to the flow cells overheating) MinKNOW has a default timeframe for the flow cell to reach the target temperature. Once the timeframe is exceeded, an error message will appear and the sequencing experiment will continue. However, sequencing at an incorrect temperature may lead to a decrease in throughput and lower q-scores. Please adjust the location of the sequencing device to ensure that it is placed at room temperature with good ventilation, then re-start the process in MinKNOW. Please refer to this link for more information on MinION temperature control. -
Guppy – no input .fast5 was found or basecalled
Observation Possible cause Comments and actions No input .fast5 was found or basecalled input_path did not point to the .fast5 file location The --input_path has to be followed by the full file path to the .fast5 files to be basecalled, and the location has to be accessible either locally or remotely through SSH. No input .fast5 was found or basecalled The .fast5 files were in a subfolder at the input_path location To allow Guppy to look into subfolders, add the --recursive flag to the command -
Guppy – no Pass or Fail folders were generated after basecalling
Observation Possible cause Comments and actions No Pass or Fail folders were generated after basecalling The --qscore_filtering flag was not included in the command The --qscore_filtering flag enables filtering of reads into Pass and Fail folders inside the output folder, based on their strand q-score. When performing live basecalling in MinKNOW, a q-score of 7 (corresponding to a basecall accuracy of ~80%) is used to separate reads into Pass and Fail folders. -
Guppy – unusually slow processing on a GPU computer
Observation Possible cause Comments and actions Unusually slow processing on a GPU computer The --device flag wasn't included in the command The --device flag specifies a GPU device to use for accelerate basecalling. If not included in the command, GPU will not be used. GPUs are counted from zero. An example is --device cuda:0 cuda:1, when 2 GPUs are specified to use by the Guppy command.
Become a full member
Purchase a MinION Starter Pack from Avantor to get full community access and benefit from:
- News - hear about the latest product updates
- Posts - interact with thousands of nanopore users from around the globe
- Software - download the latest sequencing and analysis software
Already have a Nanopore Community account?
Log in hereNeed more help?
Request a call with our experts for detailed advice on implementing nanopore sequencing.
Request a callInterested in microbiology?
Visit our microbial sequencing spotlight page on vwr.com.
Microbial sequencing









 In the image above, Sample 1 is of high molecular weight, whereas Sample 2 has been fragmented.
In the image above, Sample 1 is of high molecular weight, whereas Sample 2 has been fragmented. The pore activity plot above shows an increasing proportion of "unavailable" pores over time.
The pore activity plot above shows an increasing proportion of "unavailable" pores over time.