Welcome to the Nanopore Community
Order MinION devices and consumables
Visit vwr.comRapid barcoding DNA V14 – automated ElysION (SQK-RBK114.96)
Version for device: MinION
Introduction to the protocol
Overview of the protocol
Overview of the protocol
-
Introduction to the automated rapid barcoding DNA protocol using ElysION
The ElysION automated Rapid Barcoding Kit 96 V14 (SQK-RBK114.96) workflow is optimised for simplicity and speed to automatically prepare and sequence between 1 and 96 purified DNA input samples, then automatically wash the used flow cell for re-use. The workflow is PCR-free, removing the PCR bias and retains information about base modifications, which can be analysed using tools developed in the Nanopore Community.
Note: This kit is able to generate high sequencing accuracies of over 99% (Q20+) and duplex data; but has been optimised for speed and simplicity. If users require high accuracy, we recommend using our Native Barcoding Kit 96 V14 kit (SQK-NBD114.96).
Due to the simple nature of the workflow and the fact that little sample manipulation is required (e.g. minimal pipetting steps and no clean-up steps) long reads can be achieved with this kit, despite the required transposase fragmentation. However, in order for long reads to be observed in sequencing, long fragments need to be present in the initial sample input.
Steps in the sequencing workflow:
Prepare for your experiment
You will need to:
- Ensure your ElysION device is installed with the required hardware and software package for this workflow.
- Ensure you have your sequencing kit, the correct equipment and third-party reagents.
- Check your flow cell to ensure it has enough pores for a good sequencing run.
Automated library preparation, sequencing and analysis
The table below is an overview of the steps automated by the ElysION device.
Library preparation step Process DNA barcoding Tagmentation of the DNA using the Rapid Barcoding Kit V14 Sample pooling and clean-up Pooling of barcoded libraries and AMPure XP Bead clean-up Adapter ligation Attach the sequencing adapters to the DNA ends Priming and loading the flow cell Prime the flow cell and load the prepared library for sequencing Sequencing Your sequencing run uses the Gourami software, which will collect raw data to basecall and demultiplex the barcoded reads. Data analysis Once sequencing is complete, we have a number downstream analysis options for your data available through EPI2ME.
Equipment and consumables
- Materials
-
- 330 ng gDNA in 30 µl per sample (concentration: 11 ng/µl)
- Rapid Barcoding Kit 96 V14 (SQK-RBK114.96)
- Flow Cell Wash Kit (EXP-WSH004)
- Consumables
-
- MinION and GridION Flow Cell
- Hard-Shell® 96-Well PCR Plates, low profile, thin wall, skirted, red/clear (Bio-Rad™, cat # HSP9611)
- Thermo Scientific™ Nunc™ 96 Well 2 ml Polypropylene DeepWell Plate (Thermo Scientific, cat # 95040452)
- ElysION Disposable Tips - 1000 µl (ELY-TIP1000)
- ElysION Disposable Tips - 50 µl (ELY-TIP0050)
- ElysION Disposable Tips Box Small (ELY-TBS01)
- ElysION Disposable Tips Box Large (ELY-TBL01)
- Reservoir Plate (Porvair, cat # 390015)
- Azenta PCR Plate Lid (Azenta, cat # 4ti-0291)
- Sarstedt Screw Cap Micro tube 2 ml, sterile (Sarstedt™, cat # 72.694.006)
- Sarstedt Screw Cap Micro tube 0.5 ml, sterile (Sarstedt™, cat # 72.730.106)
- ElysION Waste Bin (ELY-WB01)
- Bovine Serum Albumin (BSA) (50 mg/ml) (e.g Invitrogen™ UltraPure™ BSA 50 mg/ml, AM2616)
- Freshly prepared 80% ethanol in nuclease-free water
- Nuclease-free water (e.g. ThermoFisher, AM9937)
- 1.5 ml Eppendorf DNA LoBind tubes
- Equipment
-
- ElysION device with MinION integration
- MinION and GridION Flow Cell Light Shield
- Microfuge
- Vortex mixer
- Microplate centrifuge, e.g. Fisherbrand™ Mini Plate Spinner Centrifuge (Fisher Scientific, 11766427)
- Multichannel pipette and tips
- P1000 pipette and tips
- P200 pipette and tips
- P100 pipette and tips
- P20 pipette and tips
- P2 pipette and tips
- Ice bucket with ice
- Optional Equipment
-
- Qubit fluorometer (or equivalent for QC check)
-
For this protocol, 330 ng gDNA in 30 µl is required per sample as input.
The starting concentration of your sample(s) should be 11 ng/µl in 30 µl of volume.
Note: The output and, sequencing read length of extracted DNA may vary depending on sample quality and species. Please ensure you are using high-quality sample inputs.
-
Third-party reagents
Depending on the extraction protocol used, not all third-party reagents are required.
We have validated and recommend the use of all the third-party reagents used in this protocol. Alternatives have not been tested by Oxford Nanopore Technologies.
For all third-party reagents, we recommend following the manufacturer's instructions to prepare the reagents for use.
-
Input DNA
How to QC your input DNA
It is important that the input DNA meets the quantity and quality requirements. Using too little or too much DNA, or DNA of poor quality (e.g. highly fragmented or containing RNA or chemical contaminants) can affect your library preparation.
For instructions on how to perform quality control of your DNA sample, please read the Input DNA/RNA QC protocol.
Chemical contaminants
Depending on how the DNA is extracted from the raw sample, certain chemical contaminants may remain in the purified DNA, which can affect library preparation efficiency and sequencing quality. Read more about contaminants on the Contaminants page of the Community.
-
Check your flow cell
The number of pores in your flow cell will be checked by the ElysION device at the start of your assay. Flow cells should be checked within 12 weeks of purchasing for MinION/GridION/PromethION.
Oxford Nanopore Technologies will replace any flow cell with fewer than the number of pores in the table below, when the result is reported within two days of performing the flow cell check, and when the storage recommendations have been followed. To do the flow cell check, please follow the instructions on the ElysION on-screen display.
Flow cell Minimum number of active pores covered by warranty MinION/GridION Flow Cell 800 PromethION Flow Cell 5000 -
Rapid Barcoding Kit 96 V14 (SQK-RBK114.96) contents
Name Acronym Cap colour No. of vials Fill volume per vial (µl) Rapid Adapter RA Green 2 15 Adapter Buffer ADB Clear 1 100 AMPure XP Beads AXP Amber 3 1,200 Elution Buffer EB Black 1 1,500 Sequencing Buffer SB Red 1 1,700 Library Beads LIB Pink 1 1,800 Library Solution LIS White cap, pink label 1 1,800 Flow Cell Flush FCF Clear 1 15,500 Flow Cell Tether FCT Purple 2 200 Rapid Barcodes RB01-96 - 3 plates 8 µl per well This Product Contains AMPure XP Reagent Manufactured by Beckman Coulter, Inc. and can be stored at -20°C with the kit without detriment to reagent stability.
Sample sheet generation
ElysION RBK114 sample sheet setup
ElysION RBK114 sample sheet setup
-
Sample sheet information
The sample sheet assigns a barcode to each sample, allowing a user to pick a range from the 96-well plate, and for a sample ID to be tracked from input to results.
For more information on setting up a sample sheet please refer to the ElysION User Guide.
-
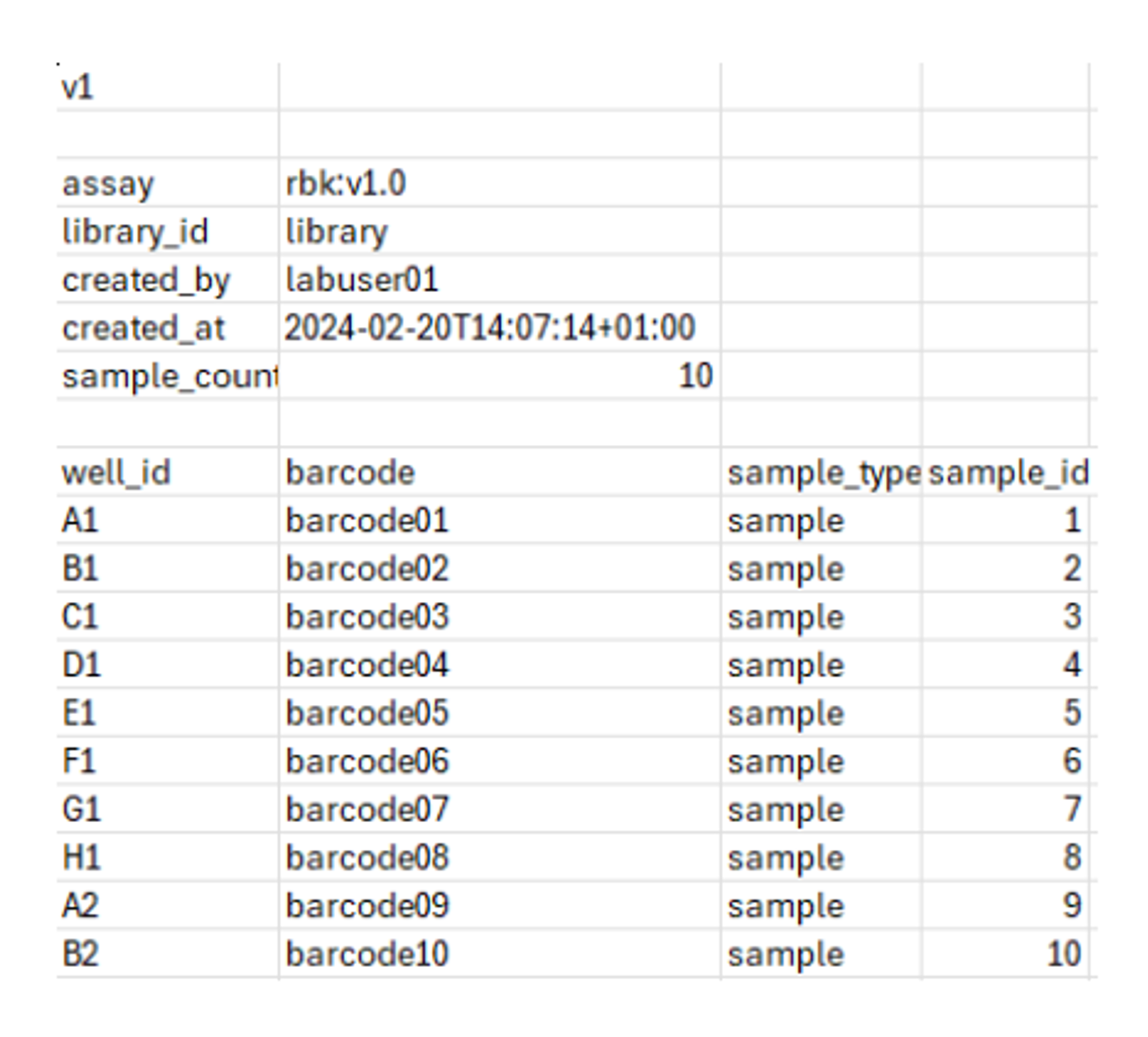
Set up your sample sheet as a .csv file with the following information:
Required field User input Definition / field info v1 Version field, no input required outside of field assay rbk:v1.0 Assay ID and version library_id User-defined Identification for the DNA library created_by User-defined Identification of operator setting up sample sheet created_at User-defined Date and time of creation. Please follow the format outlined in the ElysION user guide. sample_count User-defined Number of samples processed in run (1 to 96 samples) well_id A1-H12 Positions in 96-well plate being used (1 to 96 samples).
Samples must start from position A1 in the 96-well plate and run consecutively column-wise.barcode barcode01-barcode96 Rapid barcodes from SQK-RBK114.96 being used in library preparation.
Up to 96 barcodes are available in the sequencing kit, these should be used secuentially. (e.g. Barcodes 01-96, Barcodes 01-24, or Barcodes 50-60)sample_type User-defined Description or characteristics of sample input sample_id User-defined from LIMS system (lims-sampleid-12345) Identification for each sample input (e.g. from LIMS system)
Below is an example of a sample sheet CSV file for 10 samples:
Automated library preparation and sequencing
ElysION run setup
- Materials
-
- 330 ng gDNA in 30 µl per sample (concentration: 11 ng/µl)
- Rapid Barcode Plate (RB01-96)
- Rapid Adapter (RA)
- Adapter Buffer (ADB)
- AMPure XP Beads (AXP)
- Elution Buffer (EB)
- Consumables
-
- Hard-Shell® 96-Well PCR Plates, low profile, thin wall, skirted, red/clear (Bio-Rad™, cat # HSP9611)
- Thermo Scientific™ Nunc™ 96 Well 2 ml Polypropylene DeepWell Plate (Thermo Scientific, cat # 95040452)
- ElysION Disposable Tips - 1000 µl (ELY-TIP1000)
- ElysION Disposable Tips - 50 µl (ELY-TIP0050)
- ElysION Disposable Tips Box Small (ELY-TBS01)
- ElysION Disposable Tips Box Large (ELY-TBL01)
- Reservoir Plate (Porvair, cat # 390015)
- Azenta PCR Plate Lid (Azenta, cat # 4ti-0291)
- Sarstedt Screw Cap Micro tube 2 ml, sterile (Sarstedt™, cat # 72.694.006)
- Sarstedt Screw Cap Micro tube 0.5 ml, sterile (Sarstedt™, cat # 72.730.106)
- ElysION Waste Bin (ELY-WB01)
- Freshly prepared 80% ethanol in nuclease-free water
- Nuclease-free water (e.g. ThermoFisher, AM9937)
- Bovine Serum Albumin (BSA) (50 mg/ml) (e.g Invitrogen™ UltraPure™ BSA 50 mg/ml, AM2616)
- 1.5 ml Eppendorf DNA LoBind tubes
- Equipment
-
- Microfuge
- Vortex mixer
- Microplate centrifuge, e.g. Fisherbrand™ Mini Plate Spinner Centrifuge (Fisher Scientific, 11766427)
- P1000 pipette and tips
- P200 pipette and tips
- P100 pipette and tips
- P20 pipette and tips
- P10 pipette and tips
- P2 pipette and tips
- Multichannel pipette and tips
- Ice bucket with ice
-
The automated sample extraction, library preparation and sequencing method using the ElysION device can be followed using the on-screen display on the device.
Please ensure you have all the correct hardware components, software packages and workflows installed to carry out this method.
For more information on the device and the processes please refer to the ElysION User Guide.
-
Thaw kit components at room temperature, spin down briefly using a microfuge and mix by pipetting as indicated by the table below:
Reagent 1. Thaw at room temperature 2. Briefly spin down 3. Mix well by pipetting Rapid Barcode Plate (RB01-96) Not frozen ✓ ✓ Rapid Adapter (RA) Not frozen ✓ ✓ AMPure XP Beads (AXP) ✓ ✓ Mix by pipetting or vortexing immediately before use Elution Buffer (EB) ✓ ✓ ✓ Adapter Buffer (ADB) ✓ ✓ Mix by vortexing Flow Cell Flush (FCF) ✓ ✓ ✓ Flow Cell Tether (FCT) ✓ ✓ ✓ Sequencing Buffer (SB) ✓ ✓ ✓ Library Beads (LIB) ✓ ✓ Mix by pipetting or vortexing immediately before use Bovine Serum Albumin (BSA) ✓ ✓ ✓ Wash Mix (WMX) ✓ ✓ ✓ Wash Diluent (DIL) ✓ ✓ Mix by vortexing Storage Buffer (S) ✓ ✓ Mix by vortexing -
Switch on the ElysION device and its computer following the user guide.
-
Ensure the Background Services application is running.
If the application is not running, double click the "Background Services" desktop application on your ElysION device.
-
Open the ElysION UI application.
-
Close the door to the robot and select "Initialise" on the display.
Note: The ElysION device will perform automated checks.
Allow the initialisation checks to complete before proceeding with the library preparation method.
-
Select "Run method" on the on-screen display.
-
Select the method “Rapid Barcoding Kit” on the on-screen display.
Click next to proceed.
-
Insert a MinION and GridION Flow Cell into the MinION Mk1D device on the ElysION deck.
Instructions for inserting a flow cell onto the ElysION device can be found in the ElysION User Guide.
-
Select "Check flow cell" on the on-screen display.
Once flow cell check begins, click next to proceed.
-
Select "Import" on the on-screen display to upload your sample sheet.
Once completed click "next" to proceed.
-
In a new 0.5 ml Sarstedt Screw tube, prepare the Rapid Adapter Master Mix as follows and pipette mix:
Reagent Volume Rapid Adapter (RA) 1.5 μl Adapter Buffer (ADB) 3.5 μl Total 5 μl -
Prepare the remaining reagents in accordance with the reagent preparation page on the on-screen display.
Once completed click "next" to proceed.
-
Once your flow cell check has successfully completed, set up your sequencing run conditions on the on-screen display:
- Select the sequencing time limit, or to stop run when the flow cell pores are depleted.
- Select a data target.
- Select whether to perform:
3.1. A flow cell flush to return the used flow cell.
3.2. A flow cell wash using the Flow cell wash kit (EXP-WSH004) to reuse your flow cell.
Note: Reagent volumes and guidance for flow cell wash or flush will be given on the deck loading page. If you would like to perform a flow cell wash for re-use it is recommended to thaw the wash reagents at the same time as the library preparation reagents.
For more information on the run conditions please refer to the ElysION User Guide.
-
Set up the ElysION deck according to the deck layout outlined in the UI of on-screen display.
Note: The position of the tip boxes and reagents will change depending on the sample count, and run setup. Please ensure you are following the instructions on the on-screen display correctly for your run.
-
Insert two empty waste bins into the disposable waste draw below the deck.
-
Close the door of the ElysION device.
-
Click "Begin run" to start the automated library preparation and sequencing.
Troubleshooting
Issues during the automated library preparation
Issues during the automated library preparation
-
ElysION device troubleshooting
For commonly encountered issues please refer to the ElysION User Guide.
For in-depth troubleshooting please refer to the Set-up and operating manual: Early access ElysION provided with your ElysION device.
For additional customer support contact the nanoporetech support channels.
Become a full member
Purchase a MinION Starter Pack from Avantor to get full community access and benefit from:
- News - hear about the latest product updates
- Posts - interact with thousands of nanopore users from around the globe
- Software - download the latest sequencing and analysis software
Already have a Nanopore Community account?
Log in hereNeed more help?
Request a call with our experts for detailed advice on implementing nanopore sequencing.
Request a callInterested in microbiology?
Visit our microbial sequencing spotlight page on vwr.com.
Microbial sequencing
