Welcome to the Nanopore Community
Order MinION devices and consumables
Visit vwr.comMinION Mk1B user manual
Version for device:
Other available device versions: MinION
Overview
Device information
Device information
-
The MinION Mk1B
The Oxford Nanopore Technologies® MinION™ is an electronic device that provides the interface between the user’s computer and the nanopore sensor array. The MinION powers to the application-specific integrated circuit (ASIC), performs temperature control and transfers data to the PC through a single USB 3.0 port. The MinION Mk1B can be used with MinION flow cells and Flongle adapter and flow cells. The MinION device can be used for DNA and RNA sequencing. The MinION is operated using the MinKNOW™ software, which controls the device, experimental scripts and also performs basecalling.
-
The hardware
Component Specification Size and weight H 23 mm x W 33 mm x D 105 mm, 100 g Power 5 W Ports 1 x USB Type-B Environmental conditions Designed to sequence at 18°C to 25°C -
MinION Mk1B information
MinION Mk1B IT requirements
MinION Mk1B technical specification
Safety and regulatory information
-
FAQs
Our MinION Mk1B FAQs are located here.
Installation
What's in the box
What's in the box
-
What's in the box
The MinION Mk1B is shipped with:
- Configuration Test Cell (CTC)
- USB cable
- Quick Start Guide
Flongle adapters are shipped with a Flongle CTC.


-
The components of the MinION Mk1B are shown below.
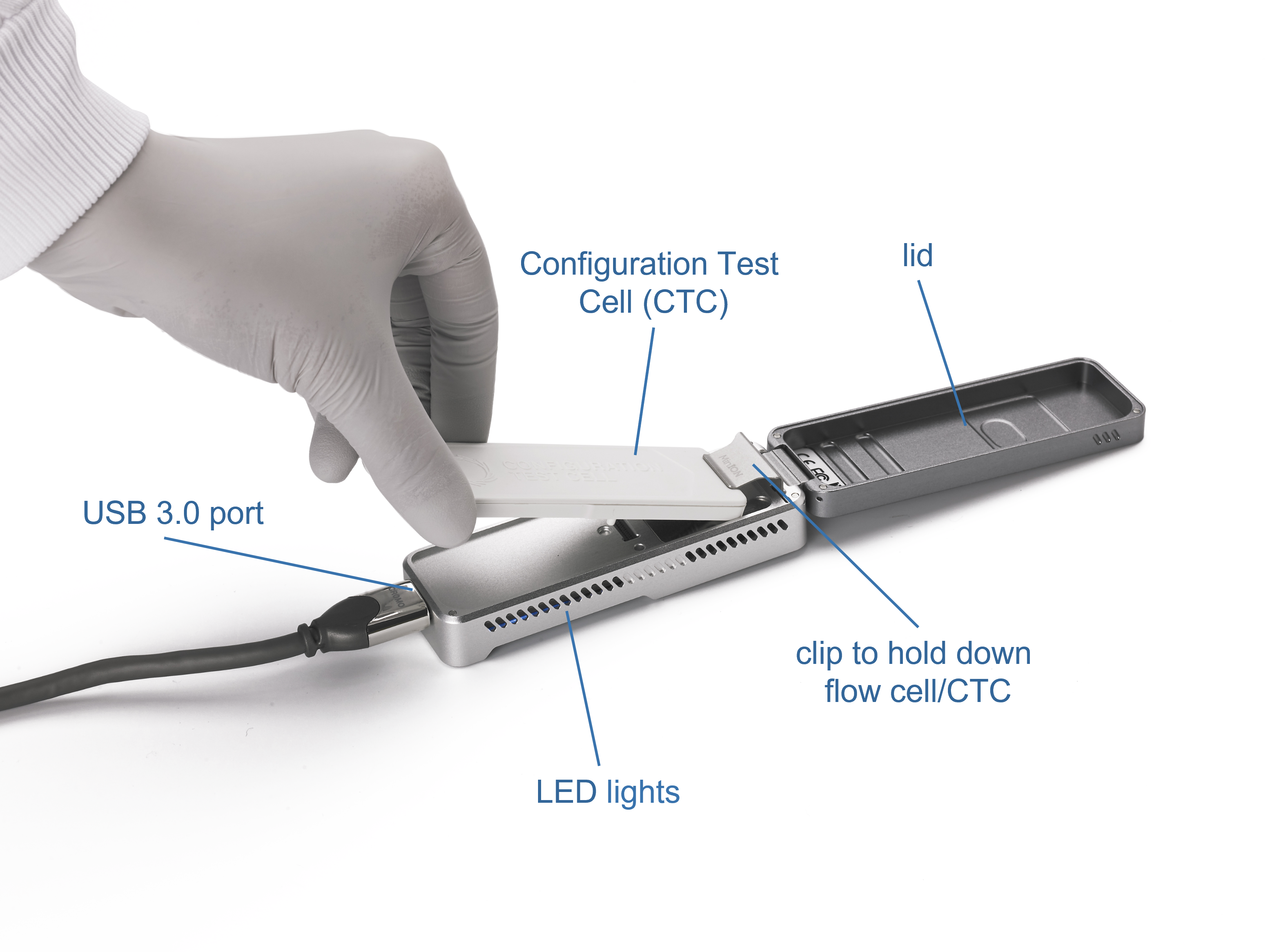
LED lights
There are four LED lights on each side of the device. The LEDs on the left side of the device display the following patterns:- (top) red LED: ON when a flow cell has been detected
- blue LED: ON when a flow cell has not been detected
- green LED: BLINKING when the flow cell has been inserted and firmware loaded
- second blue LED: BLINKING when the flow cell has been inserted and firmware loaded
The LEDs on the right side of the device display the following patterns:
- top red LED: ON when a sequencing script is running
- second red LED: ON when a flow cell has been detected
- third red LED: BLINKING when the MinKNOW software passes instructions to the MinION
- bottom red LED: BLINKING when the MinKNOW software passes instructions to the MinION
Lid
The lid can be flipped open to access the flow cell housing.
Clip
The clip holds the flow cell or Configuration Test Cell securely in place.
Configuration Test Cell (CTC)
The CTC is used during the hardware check to ensure that the communication between the device and the flow cell is working correctly.
USB 3.0 port
The USB 3.0 port is for connecting the MinION to the host computer for powering the device and data transfer.
Installing sequencing software
Installing sequencing software
-
Downloading and installing MinKNOW
All Oxford Nanopore devices use MinKNOW™ as the primary software. The MinKNOW software carries out several core tasks: data acquisition, real-time analysis and feedback, basecalling, data streaming, controlling the device, and ensuring that the platform chemistry is performing correctly to run the samples. MinKNOW takes the raw data and converts it into reads by recognition of the distinctive change in current that occurs when a DNA strand enters and leaves the pore. MinKNOW then basecalls the reads, and writes out the data into FASTQ or BAM files.
Please follow the MinKNOW protocol for information on how to install the software on Windows, Mac OS X or Linux.
Setting up the MinION Mk1B
Setting up the MinION Mk1B
- Equipment
-
- MinION Mk1B
- Configuration test cell
-
Insert the CTC into the MinION Mk1B.
- Clip the CTC into place in the MinION Mk1B and connect to the host computer.

- Gently press down on the CTC; there will be a slight click as the CTC clips into place.
- Close the MinION Mk1B lid.
-
Close the MinION Mk1B lid and connect the MinION Mk1B device to the host computer using the included USB 3.0 cable.
Setting up the MinION Mk1B for Flongle
Setting up the MinION Mk1B for Flongle
- Equipment
-
- MinION Mk1B
- Flongle adapter
-
Place the Flongle adapter into the MinION Mk1B.
The adapter should sit evenly and flat on the MinION Mk1B. This ensures the flow cell assembly is flat during the next stage.

-
Close the MinION Mk1B lid and connect the MinION Mk1B device to the host computer using the included USB 3.0 cable.
Hardware check
-
Select the sequencing device connected to the computer.
-
Click 'Start' for the check to begin.
Ensure the position selected is ready to start, as indicated on the GUI.
Using MinION flow cells:
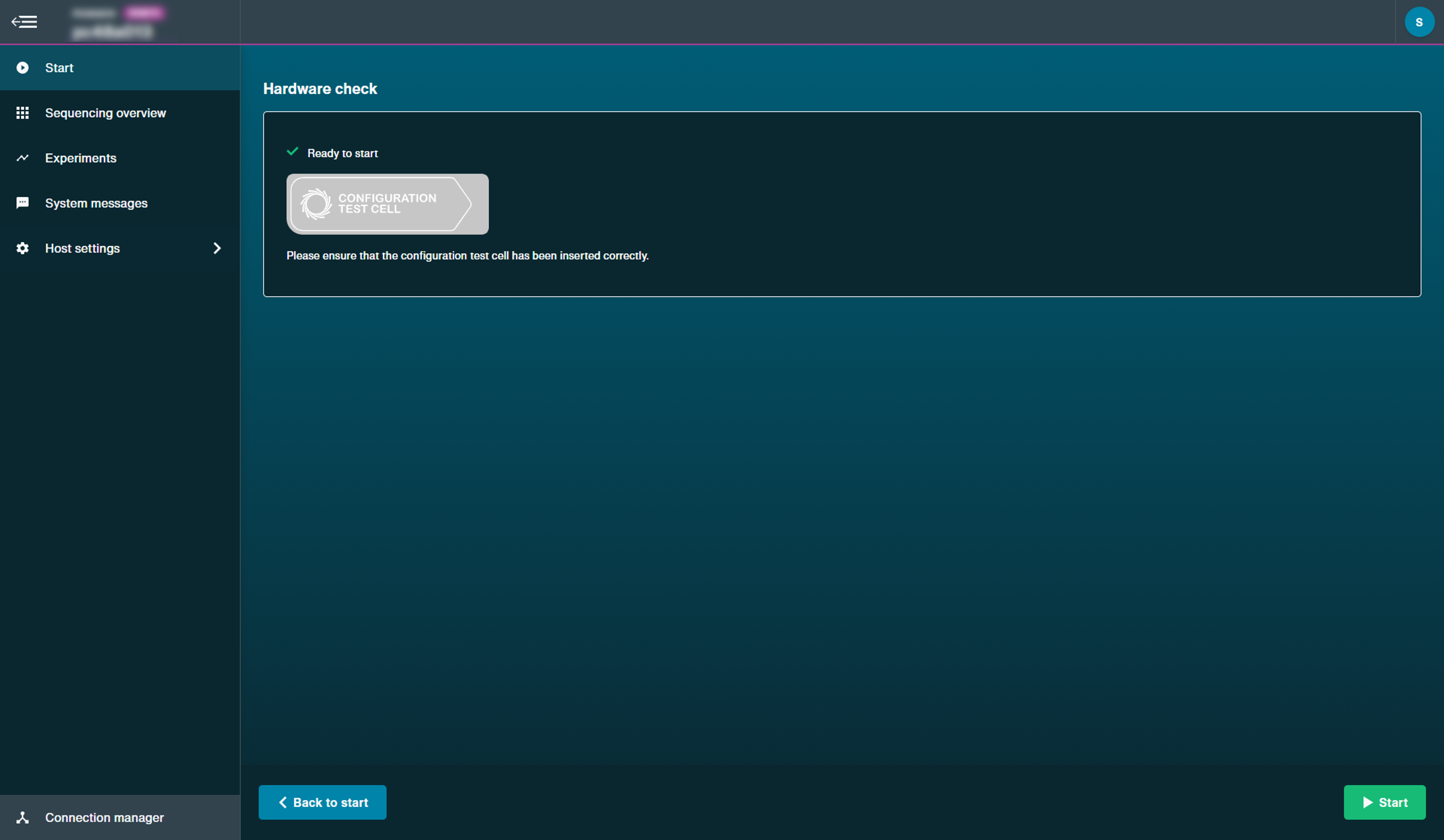
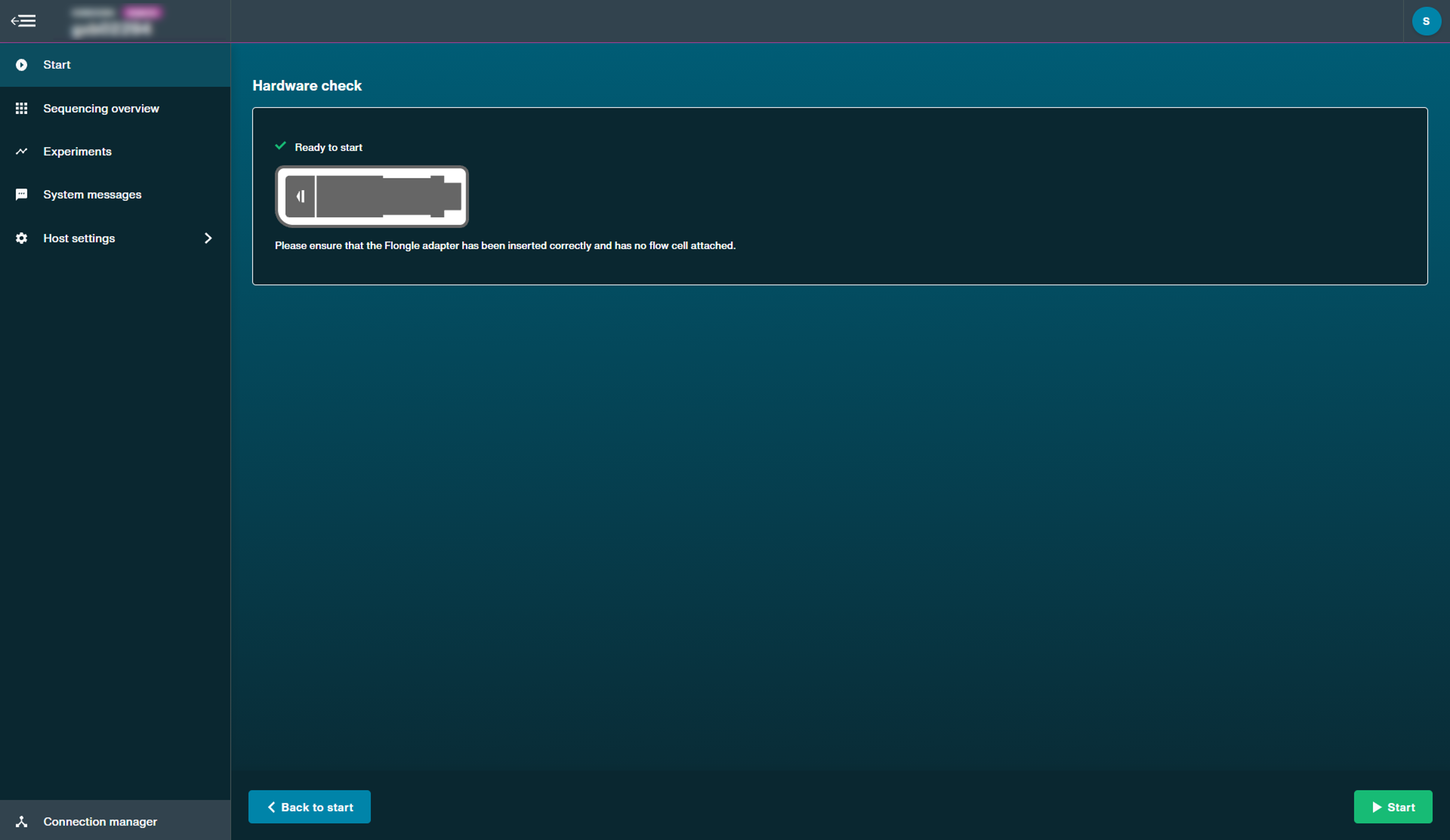
Using Flongle flow cells:
Note: To check the Flongle adapter, insert the EMPTY adapter. To check device or position, insert only a MinION CTC.
Pre-sequencing checks and sequencing run
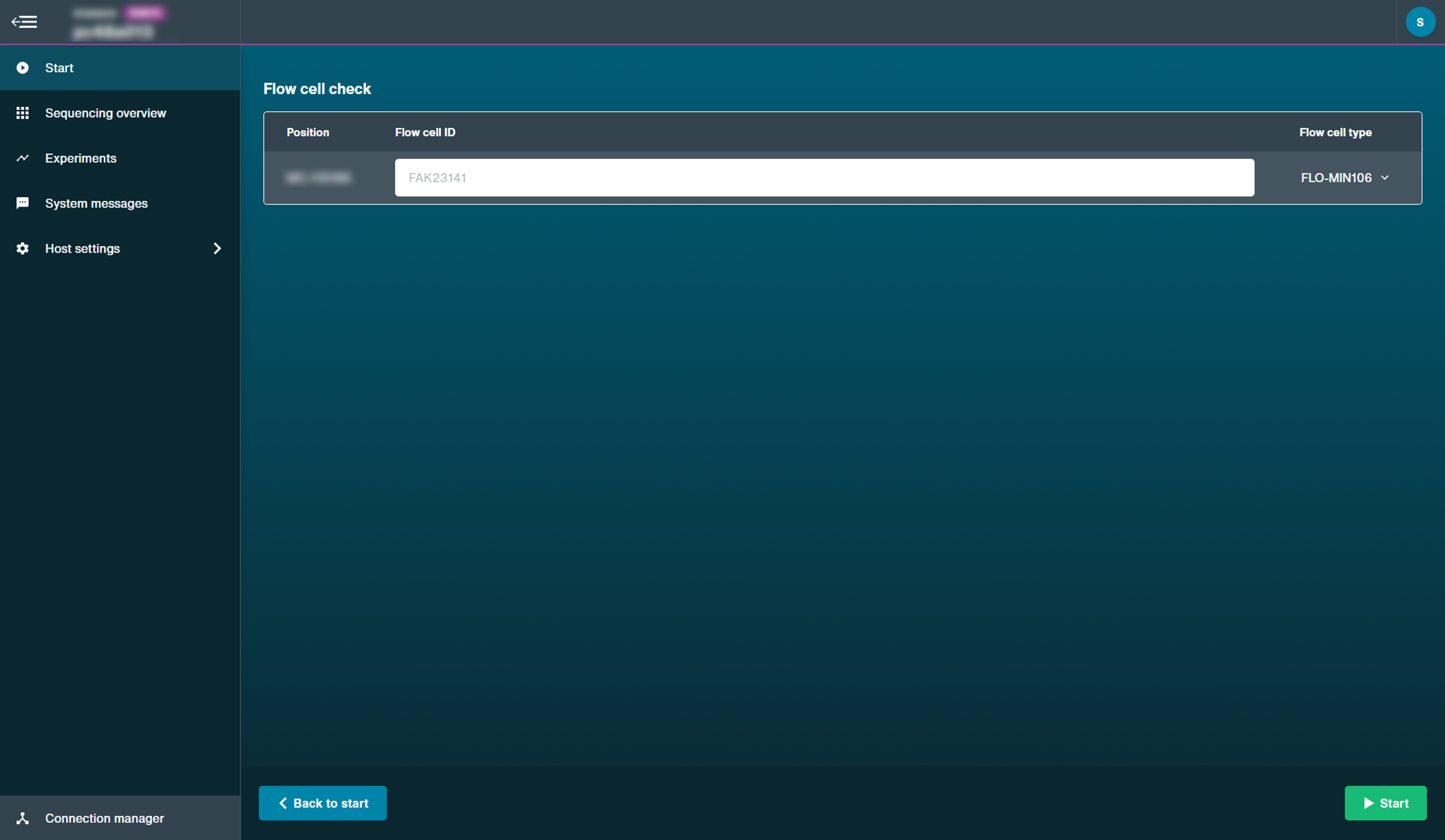
Flow cell check
Flow cell check
-
Flow cell check
We strongly recommend performing a flow cell check before loading a DNA or RNA library to assess the number of pores available.
Oxford Nanopore Technologies will replace any flow cell that falls below the warranty number of active pores within 12 weeks of purchase, provided that you report the results within two days of performing the flow cell check and you have followed the storage recommendations.
Flow cell Minimum number of active pores covered by warranty Flongle Flow Cell (FLO-FLG001) 50 MinION/GridION Flow Cell 800 PromethION Flow Cell 5000 -
When you see the MinION Flow Cell type and flow cell IDs recognised has been recognised, click 'Start' to begin.
Note: For Flongle, ensure to fill in the flow cell ID.
-
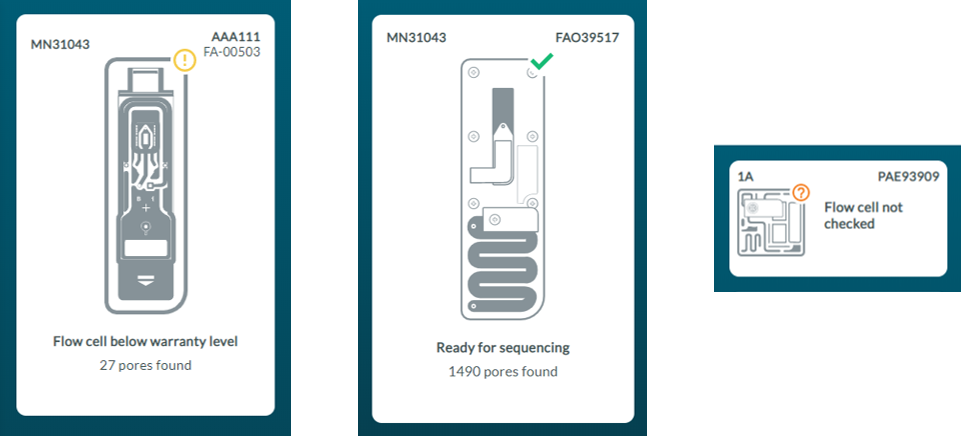
Flow cell health indicators
The quality of the flow cell will be shown as one of the three outcomes:
Yellow exclamation mark (Flongle flow cell): The number of sequencing pores is below warranty. Take the flow cell out of the device, re-insert it and run a flow cell check again. If the flow cell is still below warranty, contact support@nanoporetech.com
Green tick (MinION flow cell): The number of sequencing pores is above warranty.
Question mark (PromethION flow cell): A flow cell check has not been run on the flow cell during this MinKNOW session.
Note: The indicator of quality will only remain visible during a MinKNOW session. Once the MinKNOW session has ended, the status of the flow cell will be erased.
Sequencing and monitoring the experiment
Sequencing and monitoring the experiment
-
For instructions on how to set up a sequencing experiment and how to monitor the progress of the experiment, refer to the MinKNOW protocol in the Nanopore Community.
To run a sequencing experiment, you will need your DNA or RNA sample purified and a sequencing library prepared. For more information about sample preparation, refer to the Prepare section of our Documentation.
You can access the MinKNOW protocol by following this link.
Updating the software
MinKNOW updates for Windows and Mac OS X
-
MinKNOW updates
Availability of updates to the MinKNOW software are indicated via the MinKNOW host settings or as a pop-up when first opening the software. The user should follow the on-screen instructions to install the new versions.
The details of the update will be communicated in Nanopore Community announcements.
We strongly recommend users to update as soon as reasonably possible after the release has been made available.
Note: Users will not be able to update their device if connected remotely or if a run is in progress.
-
Open the MinKNOW UI via the desktop icon and the auto-updater will automatically check for updates when connected to the device.
-
A dialogue box will open when a new update is available.
Select Get Update to update the device software automatically.
Updates may be skipped. However, we recommend to update the device as soon as updates are available. Some updates will be mandatory to use the device and are unable to be skipped.
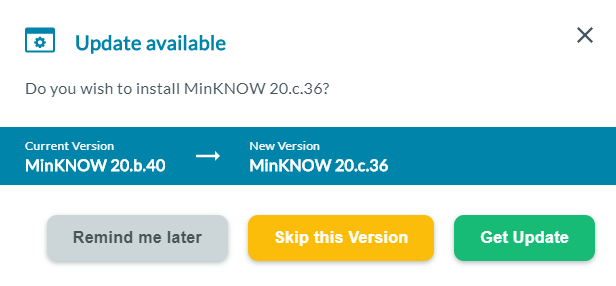
Note: For MinION Mk1B/Mk1D, clicking Get Update will open the Software Downloads page on the connected computer for you to download the updated MinION software.
-
You can also update the device from the Software page of the Host settings.
Updating MinKNOW for Linux
-
To update MinKNOW to a new version:
sudo apt clean
sudo apt update
sudo apt install ont-standalone-minknow-release
The latest version of MinKNOW will be installed. Once these commands have completed successfully without errors, reboot the device:
sudo reboot
Troubleshooting
Export logs
Export logs
-
Location of log files
Log files for each sequencing experiment can be found in:
Windows
C:\data\logsMac OS X
/private/var/log/MinKNOWLinux
/var/log/minknow
Become a full member
Purchase a MinION Starter Pack from Avantor to get full community access and benefit from:
- News - hear about the latest product updates
- Posts - interact with thousands of nanopore users from around the globe
- Software - download the latest sequencing and analysis software
Already have a Nanopore Community account?
Log in hereNeed more help?
Request a call with our experts for detailed advice on implementing nanopore sequencing.
Request a callInterested in microbiology?
Visit our microbial sequencing spotlight page on vwr.com.
Microbial sequencing














