EPI2ME Labs Nextflow Workflows demonstration
This video demonstrates running an EPI2ME Labs Nextflow workflow on the command line.
analyse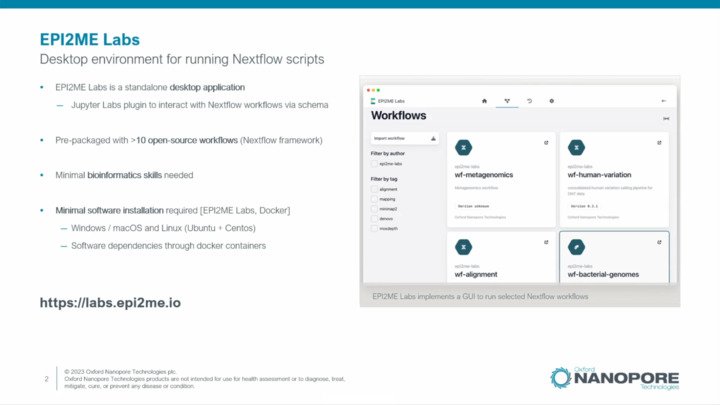
EPI2ME Labs: wf-human-variation Platform Demo
Practical demonstration on how to launch the "wf-human-variation" workflow using EPI2ME Labs Graphics User Interface.
analyse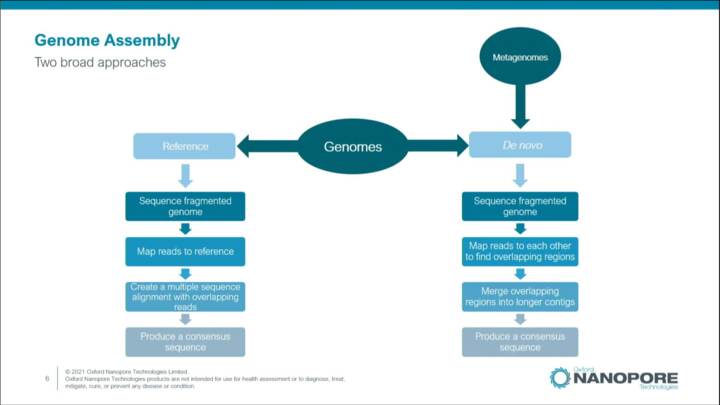
Introduction to metagenomic assembly
This video introduces concepts, tools, and techniques you can use to assemble genomes using Oxford Nanopore long read metagenomic data.
analyse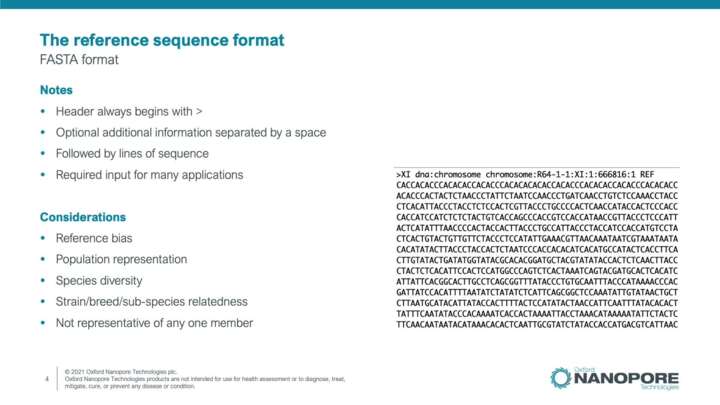
Bioinformatics file formats: reference and annotation files
This video will introduce bioinformatics file formats used to store reference sequence and annotated features data, and databases hosting these files
analyse
Introduction to read alignment/mapping
In this video you will learn about read alignment/mapping approaches, file formats and workflows
analyse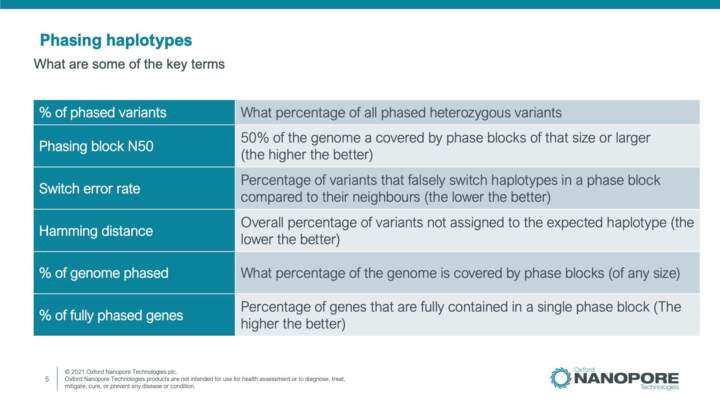
Introduction to variant annotation and phasing
In this video you will learn about tools and key terms involved in phasing genetic variation. Tools for the functional annotation of identified variants are also highlighted
analyse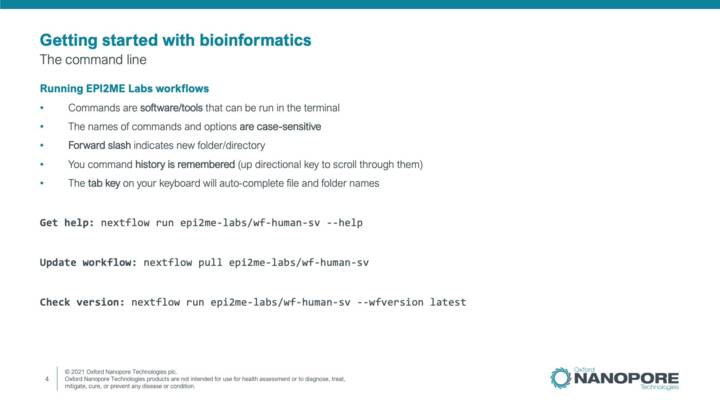
EPI2ME Labs Workflows set up and configuration
This video contains an introduction to using EPI2ME Labs workflows in the command line including commands for installing, updating and running analysis workflows. Increasing the CPU and RAM usage is also described
analyse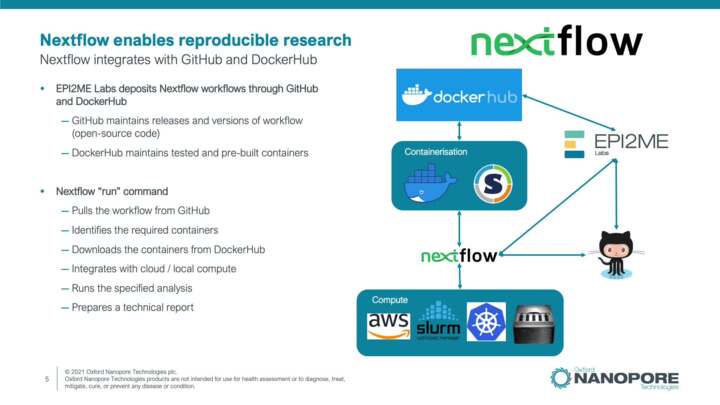
Introduction to EPI2ME Labs workflows
This video introduces bioinformatics workflows and nextflow workflows provided through the EPI2ME Labs workflows analysis platform
analyse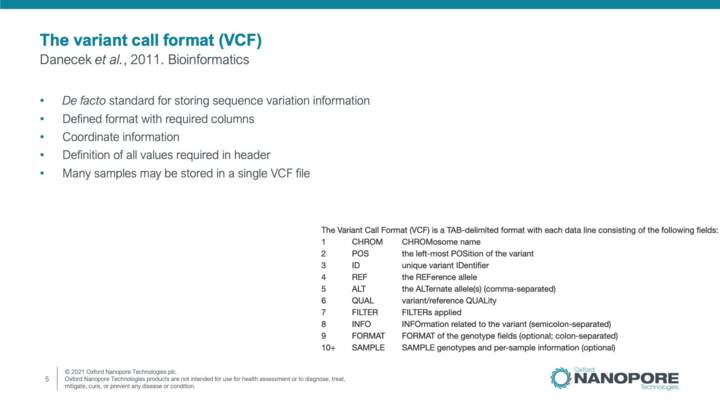
Introduction to SNP and indel detection
This video will introduce the general workflow for detection SNP and indel variation, relevant file formats and ONT analysis solutions for detection of SNPs and indels in your data
analyse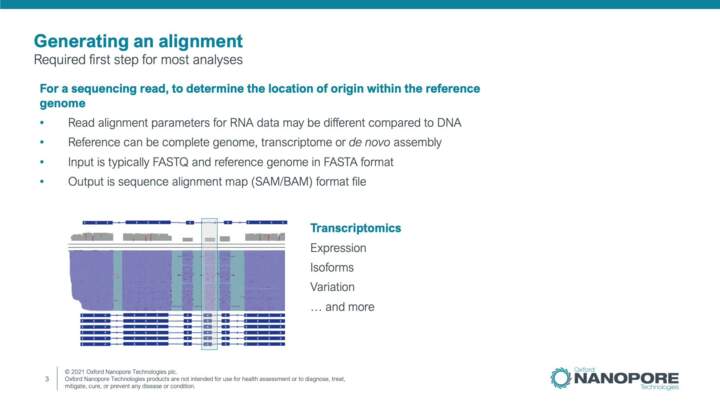
Differential gene expression analysis workflows
This video will introduce steps involved required to detect differentially expressed genes and bioinformatics solutions available to help with this process
analyse
Introduction to differential gene expression
This video will introduce an overview of the process for detection of differentially expressed genes and considerations for processing and interpreting transcriptomic data and results
analyse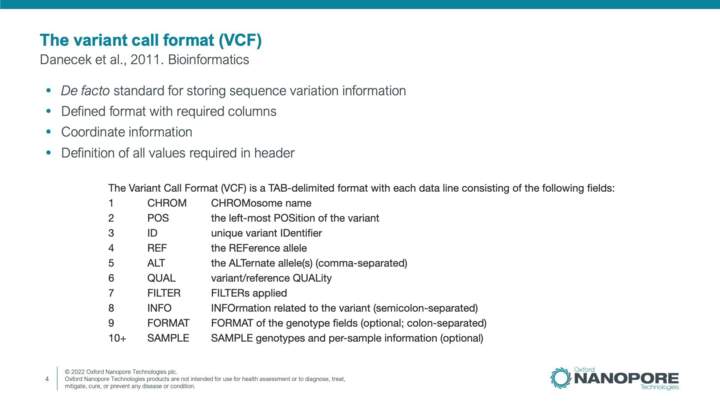
Introduction to structural variation detection
This video will explain structural variation definitions, and introduce relevant file formats and detection workflows
analyse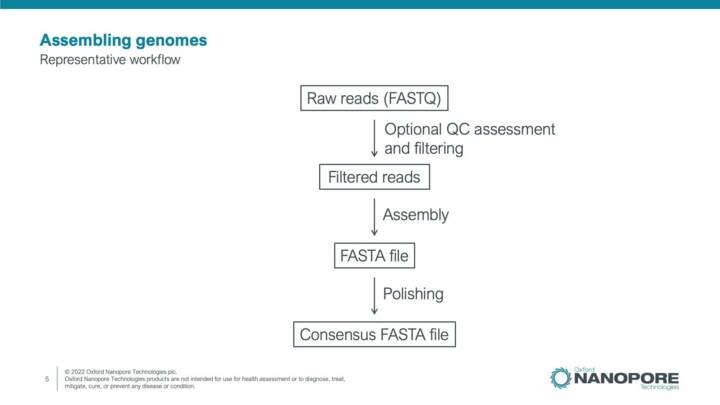
Introduction to de novo genome assembly and polishing
This video will introduce steps and workflows for the generation of high quality consensus genome sequences
analyse
Introduction to read quality assessment and filtering
In this video you will learn about tools for assessing and filtering sequencing read data
analyse
Data Analysis: EPI2ME Labs introduction
EPI2ME is a user-friendly analysis platform, highlighting best practice tools and analysis workflows for many applications. This video contains a short summary introducing the platform
analyse
Introduction to metagenomics classification concepts
This video introduces metagenomic classification and some of the key concepts necessary to help plan a successful metagenomic classification analysis workflow
analyse
Introduction to metagenomics
This video introduces the Metagenomics course and gives an overview of the flexible workflow options available to help plan and perform your metagenomics experiments.
plan analyse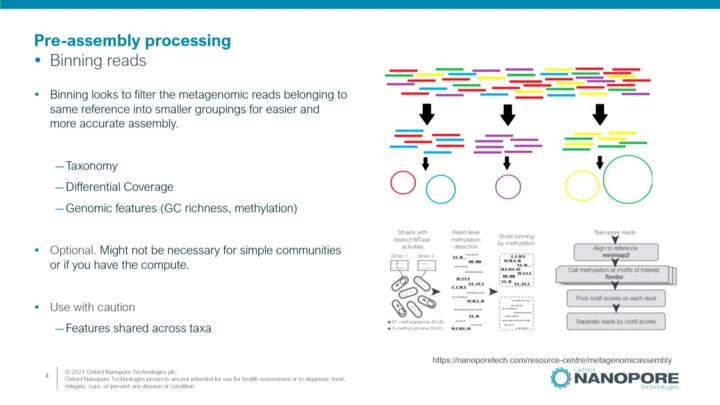
Additional concepts in metagenomic assembly
This video introduces additional concepts to help plan and perform metagenomic assembly analysis.
analyse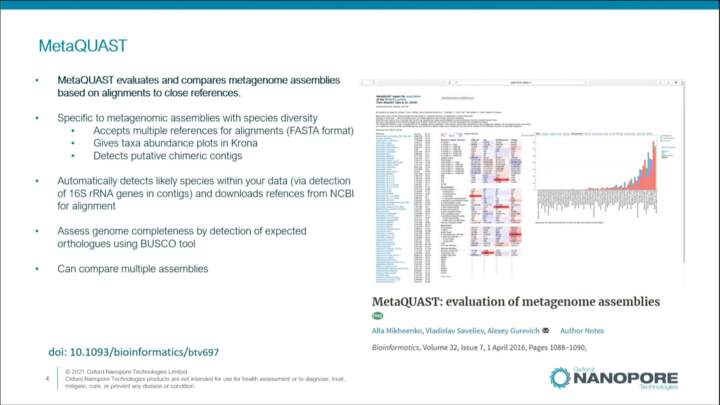
Quality control of metagenomic assemblies
An overview into the concepts and tools you can use to assess the quality of your metagenomic assemblies.
analyse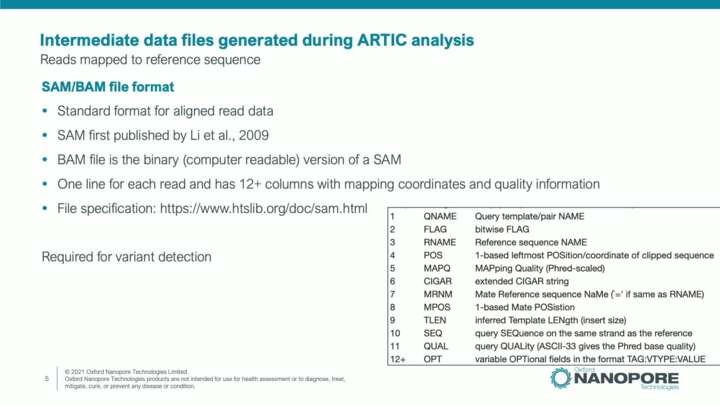
SARS-CoV-2: ARTIC workflow data and analysis files
In this video we will introduce and discuss the main file types and file formats for the data files that are required and generated by the ARTIC Field Bioinformatics analysis workflow. This will explain the FASTQ, BAM, VCF and FASTA format files.
analyse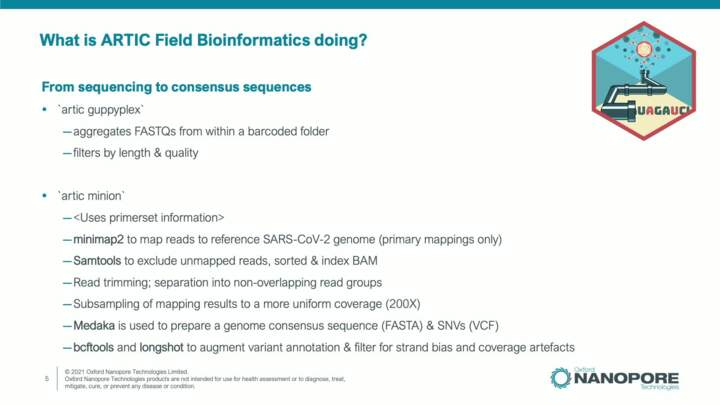
SARS-CoV-2: ARTIC Field Bioinformatics analysis technical details
The ARTIC Field Bioinformatics workflow involves several analysis steps whereby data generated from a sequencing device is used to detect variants and generate a high-confidence consensus sequence. In this video we will explain the individual components and analysis software of this workflow
analyse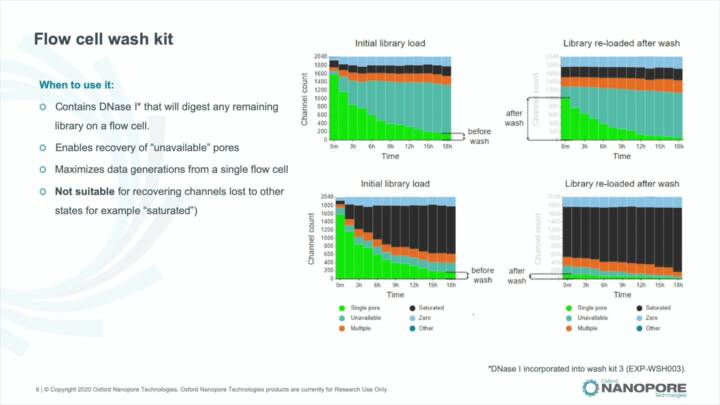
MinKNOW: Blocking
Using the MinKNOW GUI to identify and troubleshoot blocking during a sequencing run
sequence analyse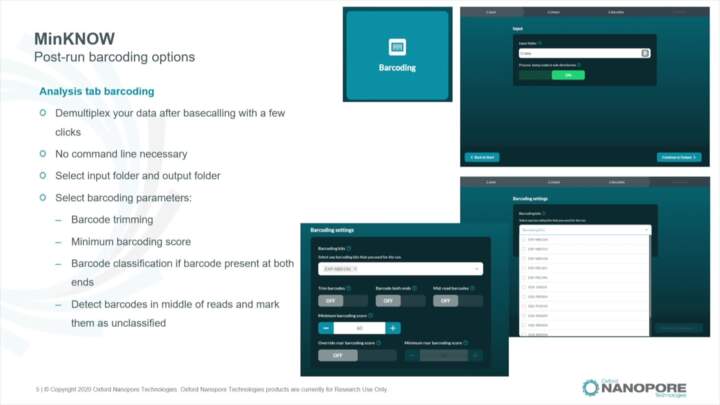
MinKNOW: Post-run analysis
Learn about the main post-run analysis processes performed by MinKNOW
sequence analyse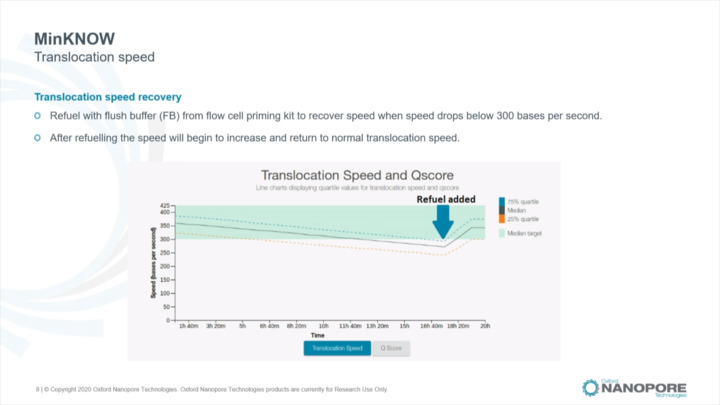
MinKNOW: Translocation speed
Understand translocation speed and how to identify and troubleshoot translocation speed changes
analyse
MinKNOW: The graphical user interface (GUI)
Guided tour through MinKNOW's graphical user interface
prepare sequence analyse
MinKNOW: Live basecalling and output folder structure
Set up live basecalling in MinKNOW and chose basecalling model. Introduction to data file types and folder structure.
sequence analyse
Knowledge Exchange: Structural variation
This Knowledge Exchange provides an overview of structural variation detection using long reads.
analyse
Knowledge Exchange: cDNA sequencing on the Oxford Nanopore platform
This Knowledge Exchange introduces cDNA sequencing with Oxford Nanopore technology - from library prep to analysis.
prepare analyse
Knowledge Exchange: Assembly
With the increasing yield of the Oxford Nanopore platform, it’s becoming easier to generate enough data to assemble large genomes. In this Knowledge Exchange, learn how to plan and perform genome assembly projects using nanopore sequencing.
analyse
Knowledge Exchange: Direct RNA sequencing
This Knowledge Exchange discusses nanopore direct RNA sequencing; a highly parallel, real-time, single-molecule method that circumvents reverse transcription or amplification steps.
prepare analyse