
Priming and loading your flow cell
Flow cell priming and sample loading demonstration.
sequence
Knowledge Exchange: GridION Mk1: delivering ultimate flexibility in the lab
This Knowledge Exchange introduces the GridION Mk1 sequencing device - capable of running up to 5 individually addressable MinION or Flongle Flow Cells.
sequence
A practical demonstration of priming and loading a Flongle flow cell
A short video describing how to prime and load a flongle flow cell.
plan prepare sequence
A practical demonstration of how to prime and load a PromethION flow cell
In this video you will learn how to prime and load a promethION flow cell
sequence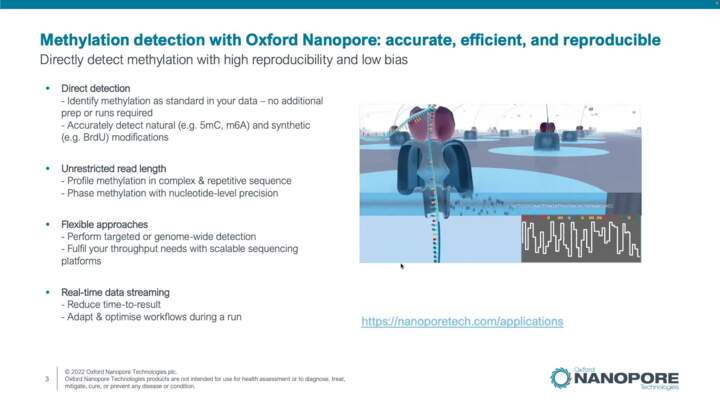
Methylation Detection: Sample to Answer Workflow Overview
This video introduces methylation detection capabilities of Nanopore sequencing and includes general recommendations for preparing your library for methylation detection.
plan prepare sequence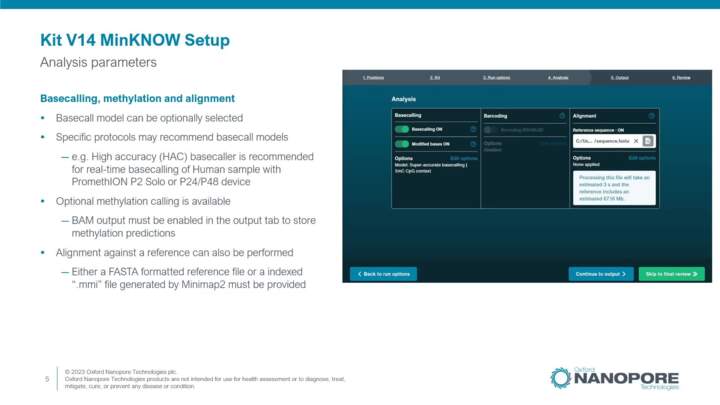
MinKNOW Configuration for Kit 14
Illustration on how to setup a Kit V14 sequencing run using MinKNOW software.
sequence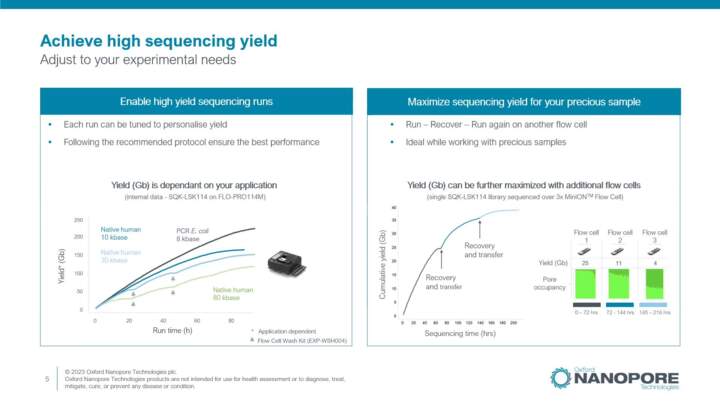
Kit V14 performance
An overview of Kit 14 performance when compared to previous chemistries.
plan prepare sequence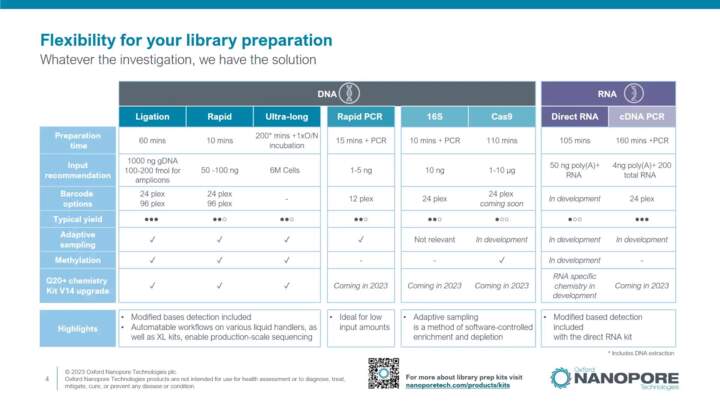
Kit V14 introduction and overview
An introduction to Kit 14 and chemistry overview.
plan sequence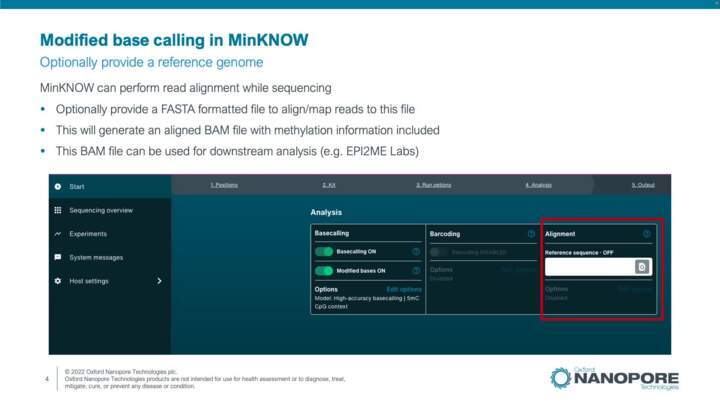
Methylation Detection On MinKNOW
This video describes tools and approaches for methylation detection, setting up a run on the MinKNOW software and tools for downstream analysis of methylation data.
plan sequence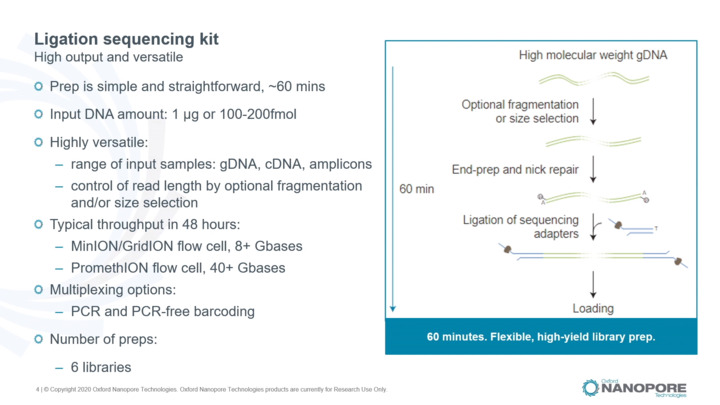
Library preparation: Ligation sequencing kit
Presenting the general workflow of the ligation sequencing kit, recommended input, options to scale-up, and the barcoding options available.
prepare sequence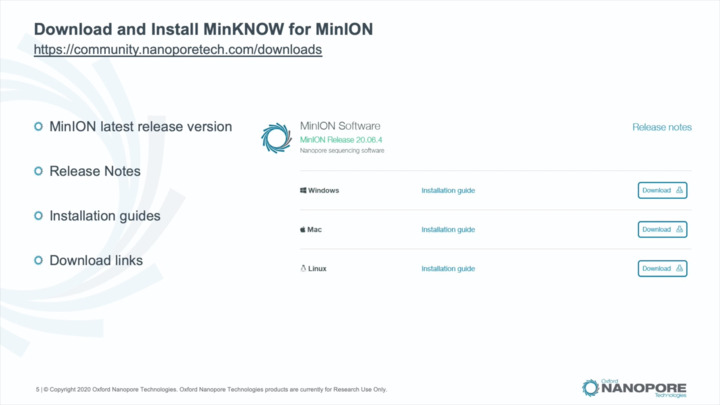
MinKNOW: Installation
Learn about software releases, downloading and installing MinKNOW, and troubleshooting installation.
plan sequence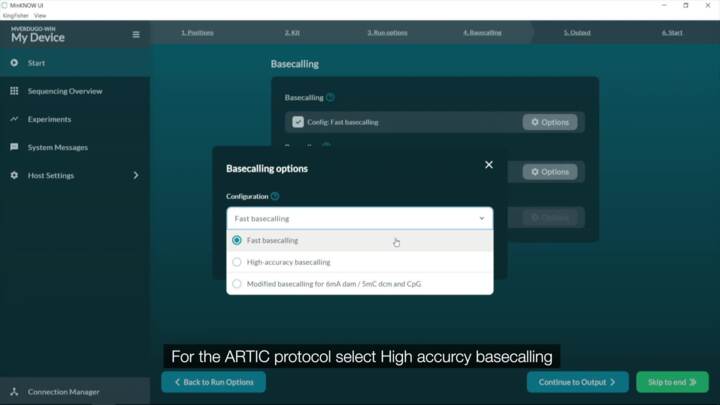
SARS-CoV-2: Setting up a new ARTIC experiment
Demonstration on setting up a new experiment on the MinKNOW software.
sequence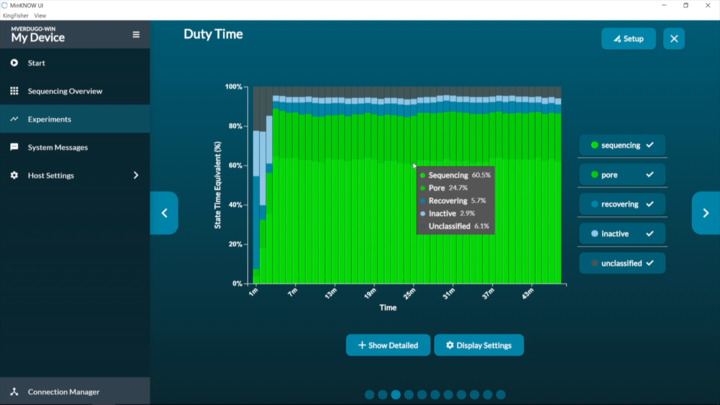
MinKNOW: Assessing your run
Tour through MinKNOW during a live run to understand the different graphs and assess the quality of your run.
sequence
A practical demonstration of priming and loading a flow cell
Flow cell priming and sample loading demonstration
sequence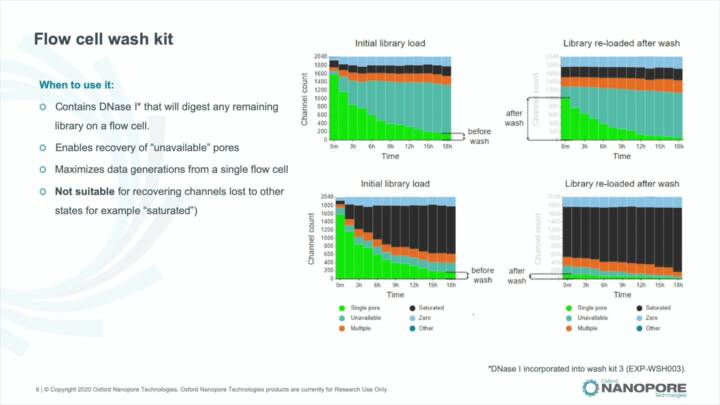
MinKNOW: Blocking
Using the MinKNOW GUI to identify and troubleshoot blocking during a sequencing run
sequence analyse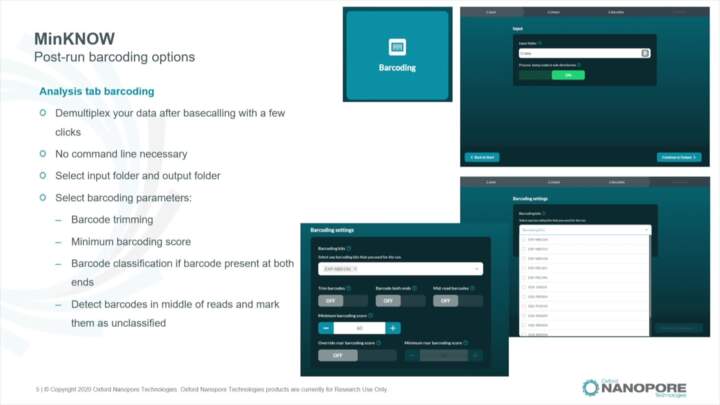
MinKNOW: Post-run analysis
Learn about the main post-run analysis processes performed by MinKNOW
sequence analyse
MinKNOW: Re-using a flow cell
The wash kit explained, how to adjust bias voltage when reusing a flow cell in MinKNOW.
sequence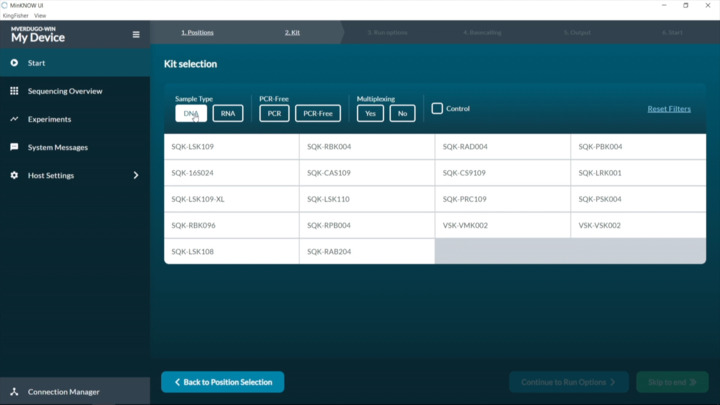
MinKNOW: Setting up a new experiment
Demonstration on setting up a new experiment on the MinKNOW software.
sequence
MinKNOW: The graphical user interface (GUI)
Guided tour through MinKNOW's graphical user interface
prepare sequence analyse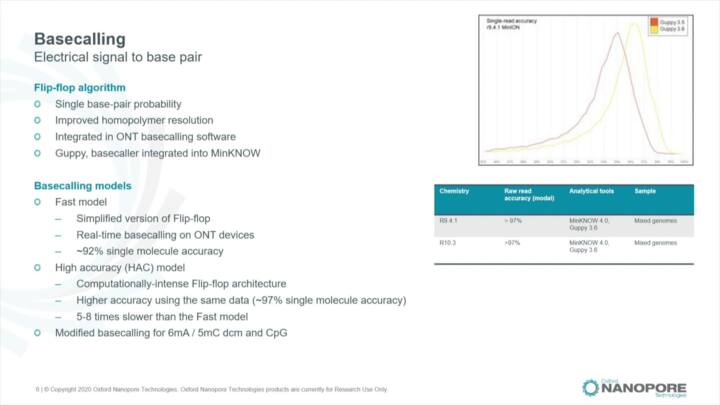
MinKNOW: Live basecalling and output folder structure
Set up live basecalling in MinKNOW and chose basecalling model. Introduction to data file types and folder structure.
sequence analyse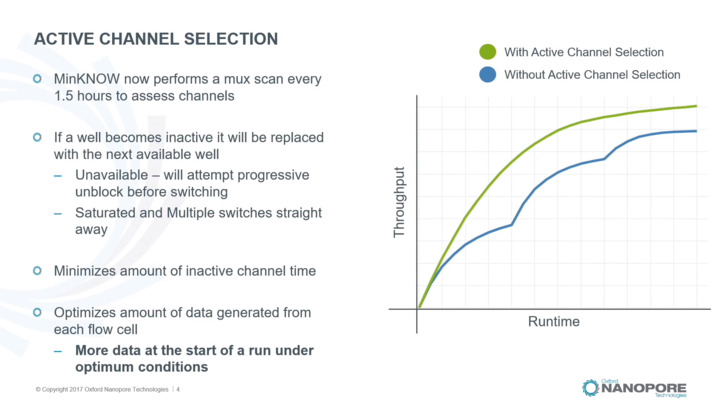
MinKNOW: Mux scan and active channel selection
Introduction to MUX scan and active channel selection.
sequence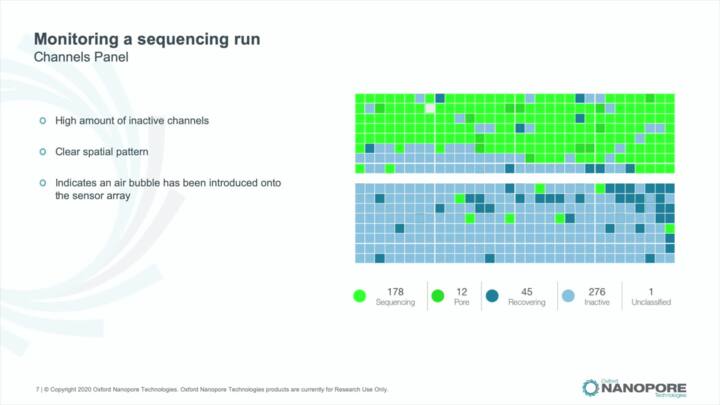
MinKNOW: Pore occupancy
Introduction to pore occupancy, how to monitor and interpret this during a run, and how to troubleshoot for different scenarios.
sequence
Knowledge Exchange: Library preparation kits and methods
This Knowledge Exchange covers the essentials of sample and library preparation for Oxford Nanopore Technologies sequencing devices.
sequence
Knowledge Exchange: Getting the most out of a flow cell
This Knowledge Exchange provides top tips for getting the most out of a nanopore sequencing flow cell.
sequence
Knowledge Exchange: MinKNOW features and updates
This Knowledge Exchange provides an overview of the latest MinKNOW features.
sequence
Knowledge Exchange: Nanopore from 100 to 1000 genomes: towards a better understanding of phenotypes
In this Knowledge Exchange, Fritz Sedlazeck discusses his work on characterising human genetic variation with nanopore sequencing.
sequence
Knowledge Exchange: No assembly required: nanopore sequencing complete virus genomes from microbial communities
In this Knowledge Exchange, John Beaulaurier discusses an assembly-free nanopore sequencing and analysis approach for direct recovery of high-quality marine phage genome sequences.
sequence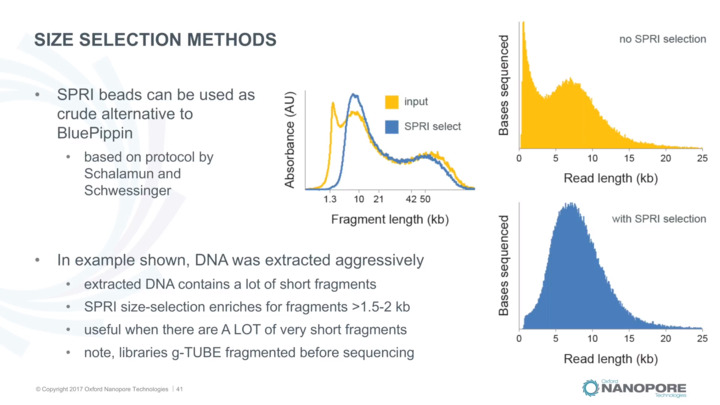
Improving sequencing yield
Getting the most out of your samples
prepare sequence
Ligation Lambda Control Experiment
Overview of Ligation Lambda Control Experiment, the library preparation and setting up a control run in MinKNOW
sequence
Rapid Lambda Control Experiment
Overview of rapid lambda control experiment
sequence